最近打算系统的回顾一下机器学习算法,所以以《机器学习实战》为依据,这次就由k-近邻算法开始回顾。
k-近邻算法概述 k-近邻算法(K-Nearest Neighbor )的工作原理是:存在一个样本数据集合,也称为训练样本集,并且样本集中每个数据都存在标签,即我们知道样本集中每一数据与所属分类对应的关系。输入没有标签的新数据后,将新数据中的每个特征与样本集中数据对应的特征进行比较,然后算法提取样本集中特征最相似数据(最近邻)的分类标签。一般来说,我们只选择样本数据集中前k个最相似的数据,这就是k-近邻算法中k的出处,通常k是不大于20的整数。最后,选择k个最相似数据中出现次数最多的分类,作为新数据的分类。
以使用k-近邻算法分类爱情片和动作片,有人统计过很多电影的打斗镜头和接吻镜头如下表,假如有一部未看过的电影,以?来表示,如何确定它是爱情片和动作片呢?
电影名称
打斗镜头
接吻镜头
电影类型
Califoria Man
3
104
爱情片
He’s Not Really into Dudes
2
100
爱情片
Beautigul Woman
1
81
爱情片
Kevin Longblade
101
10
动作片
Robo Slayer 3000
99
5
动作片
Amped II
98
2
动作片
?
18
90
未知
在KNN中,通过计算对象间距离来作为各个对象之间的非相似性指标,避免了对象之间的匹配问题,在这里距离一般使用欧氏距离或曼哈顿距离:
我们用欧氏距离来得到如下距离值:
电影名称
与未知电影的距离
Califoria Man
20.5
He’s Not Really into Dudes
18.7
Beautigul Woman
19.2
Kevin Longblade
115.3
Robo Slayer 3000
117.4
Amped II
118.9
按照距离递增排序,可以找到k个距离最近的电影。假设k=3,则三个最靠近的电影依次是He’s Not Really into Dudes、Beautigul Woman和Califoria Man。k-近邻算法按照距离最近的三部电影的类型,决定未知电影的类型,而这三部电影全是爱情片,因此我们判定未知电影是爱情片。
准备:使用Python导入数据 首先,创建名为KNN.py的python模块:
1 2 3 4 5 6 7 import numpy as npimport operatordef createDataSet () : group = np.array([[1.0 ,1.1 ],[1.0 ,1.0 ],[0 ,0 ],[0 ,0.1 ]]) labels = ['A' ,'A' ,'B' ,'B' ] return group, labels
这里我们准备了4组数据,每组数据有两个我们已知的属性或者特征值。接下来我们将完成分类任务。
实施KNN分类算法 定义函数classify0()如下:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 def classify0 (inX, dataSet, labels, k) : dataSetSize = dataSet.shape[0 ] diffMat = np.tile(inX, (dataSetSize,1 )) - dataSet sqDiffMat = diffMat**2 sqDistances = sqDiffMat.sum(axis=1 ) distances = sqDistances**0.5 sortedDistIndicies = distances.argsort() classCount={} for i in range(k): voteIlabel = labels[sortedDistIndicies[i]] classCount[voteIlabel] = classCount.get(voteIlabel,0 ) + 1 sortedClassCount = sorted(classCount.items(),key=operator.itemgetter(1 ),reverse=True ) return sortedClassCount[0 ][0 ]
至此,我们构建了第一个分类器。
示例:使用k-近邻算法改进约会网站的配对效果 海伦一直使用在线约会网站寻找自己的约会对象,她把自己交往过的对象分为以下三类:
因此,她希望更好地将她匹配的对象分类。
准备数据:从文本文件中解析数据 她把数据存放在文本文件datingTestSet2.txt中,每个样本数据占据一行,主要包含以下3种特征:
每年获得的飞行常客里程数
玩视频游戏所耗时间百分比
每周消费的冰淇淋公升数
创建名为file2matrix的函数,把数据转变为分类器可以接受的格式。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 def file2matrix (filename) : fr = open(filename) arrayOlines = fr.readlines() numberOfLines = len(arrayOlines) returnMat = np.zeros((numberOfLines,3 )) classLabelVector = [] index = 0 for line in arrayOlines: line = line.strip() listFromLine = line.split('\t' ) returnMat[index,:] = listFromLine[0 :3 ] classLabelVector.append(int(listFromLine[-1 ])) index += 1 return returnMat,classLabelVector
接下来图形化展示数据内容。
分析数据:使用Matplotlib创建散点图 定义一个scatterPlot函数进行可视化。
1 2 3 4 5 6 7 8 import matplotlibimport matplotlib.pyplot as pltdef scatterPlot () : fig = plt.figure() ax = fig.add_subplot(111 ) p = ax.scatter(datingDateMat[:,1 ], datingDateMat[:,2 ], 15.0 *np.array(datingLabels), 15.0 *np.array(datingLabels)) plt.show()
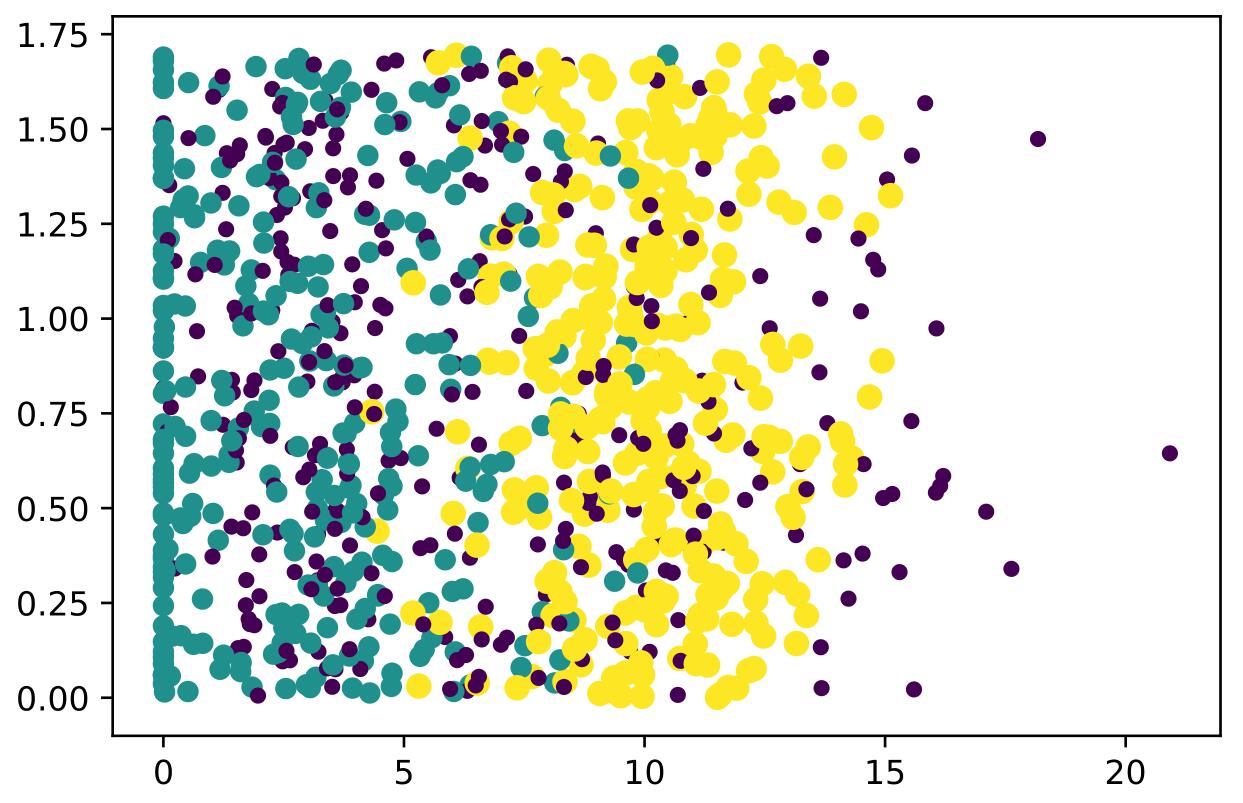
得到了玩视频游戏所耗时间百分比和每周消费的冰淇淋公升数的关系散点图:
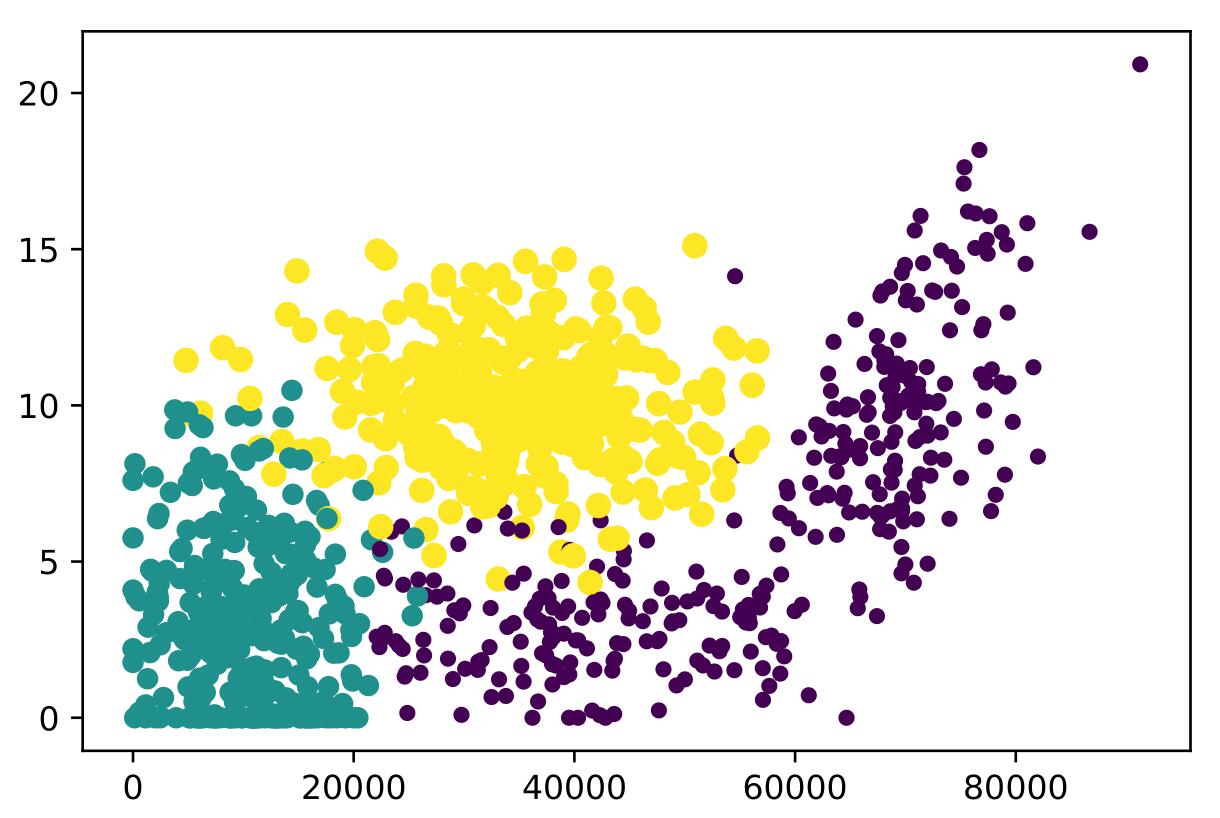
同理也能得到每年获得的飞行常客里程数和玩视频游戏所耗时间百分比的关系散点图,可以得到更好的展示效果:
可以知道具有不同的爱好的人其类别区域也不同。
准备数据:归一化数值
玩视频游戏所耗时间百分比
每年获得的飞行常客里程数
每周消费的冰淇淋公升数
样本分类
1
0.8
400
0.5
1
2
12
134000
0.9
3
3
0
20000
1.1
2
4
67
32000
0.1
2
上表给出提取的四组数据,要计算样本3和4的距离,使用以下方法:
容易发现,上面方程中数值差值最大的属性对计算结果的影响最大,也就是说,每年获取的飞行常客里程数对于计算结果的影响将远远大于该表其他两个特征的影响。而三种特征是同等重要的,因此需要把数值归一化。
增加函数autoNorm用于归一化:
1 2 3 4 5 6 7 8 9 def autoNorm (dataSet) : minVals = dataSet.min(0 ) maxVals = dataSet.max(0 ) ranges = maxVals - minVals normDateSet = np.zeros(np.shape(dataSet)) m = dataSet.shape[0 ] normDateSet = dataSet - np.tile(minVals, (m,1 )) normDateSet = normDateSet/np.tile(ranges, (m,1 )) return normDateSet, ranges, minVals
在机器学习框架scikit-learn中,有封装好的标准化函数:
1 2 3 4 5 6 7 8 9 10 11 from sklearn.preprocessing import MinMaxScalerss = MinMaxScaler() X = ss.fit_transform(X) from sklearn.preprocessing import StandardScalerss = StandardScaler() X = ss.fit_transform(X)
测试算法:作为完整程序验证分类器 创建函数datingClassTest以测试分类器效果:
1 2 3 4 5 6 7 8 9 10 11 12 def datingClassTest () : hoRatio = 0.10 datingDataMat,datingLabels = file2matrix('datingTestSet2.txt' ) normMat, ranges, minVals = autoNorm(datingDataMat) m = normMat.shape[0 ] numTestVecs = int(m*hoRatio) errorCount = 0.0 for i in range(numTestVecs): classifierResult = classify0(normMat[i,:],normMat[numTestVecs:m,:],datingLabels[numTestVecs:m],3 ) print("the classifier came back with: %d, the real answer is: %d" % (classifierResult, datingLabels[i])) if (classifierResult != datingLabels[i]): errorCount += 1.0 print("the total error rate is: %f" % (errorCount/float(numTestVecs)))
通过分类器可得错误率为5%,结果还不错,也可以改变hoRatio和k来得到不同结果。
这个例子表明我们可以得到较好的分类结果,海伦可以输入未知对象的属性,由分类软件来帮助她判定某一对象的可交往程度:讨厌、一般喜欢、非常喜欢。
示例:手写识别系统 上面的例子使用的数据比较容易理解,如何在人不太容易看懂的数据上使用分类器呢?我们将使用k-近邻分类器用于手写数字识别。
准备数据:将图像转换为测试向量 首先把图像格式化处理为一个向量。即将32×32的二进制文本。如果要使用我们的分类器,就将其变成1×1024的向量。
编写函数实现:
1 2 3 4 5 6 7 8 def img2vector (filename) : returnVect = np.zeros((1 ,1024 )) fr = open(filename) for i in range(32 ): lineStr = fr.readline() for j in range(32 ): returnVect[0 ,32 *i+j] = int(lineStr[j]) return returnVect
测试算法:使用k-近邻算法识别手写数字 使用以下代码测试分类器:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 hwLabels = [] trainingFileList = os.listdir('trainingDigits' ) m = len(trainingFileList) trainingMat = np.zeros((m,1024 )) for i in range(m): fileNameStr = trainingFileList[i] fileStr = fileNameStr.split('.' )[0 ] classNumStr = int(fileStr.split('_' )[0 ]) hwLabels.append(classNumStr) trainingMat[i,:] = img2vector('trainingDigits/%s' % fileNameStr) testFileList = os.listdir('testDigits' ) errorCount = 0.0 mTest = len(testFileList) for i in range(mTest): fileNameStr = testFileList[i] fileStr = fileNameStr.split('.' )[0 ] classNumStr = int(fileStr.split('_' )[0 ]) vectorUnderTest = img2vector('testDigits/%s' % fileNameStr) classifierResult = classify0(vectorUnderTest, trainingMat, hwLabels, 3 ) print("the classifier came back with: %d, the real answer is: %d" % (classifierResult, classNumStr)) if (classifierResult != classNumStr): errorCount += 1.0 print("\nthe total number of errors is: %d" % errorCount) print("\nthe total error rate is: %f" % (errorCount/float(mTest)))
本函数是将trainingDigits目录中的文件内容存储在列表中,然后得到目录中有多少文件,并将其存储到变量m中。接着,创建m行1024列的训练矩阵,该矩阵的每行数据存储一个图像。我们可以从文件名中解析出分类数字。该目录下的文件按照规则命名,如文件9_45.txt的分类是9,它是数字9的第45个实例。然后我们可以将类代码存储到hwLabels向量中,使用前面讨论的img2vector函数载入图像。在下一步中,我们对testDigits目录中的文件执行相似的操作,不同之处是我们并不将这个目录下的文件载入矩阵中,而是使用classify0()函数测试该目录下的每个文件。由于文件中的值已经在0和1之间,并不需要进行标准化。
通过测试本分类器,发现有相对理想的结果。
实际使用本算法时,算法的执行效率并不高。因为算法需要为每个测试向量做2000次距离计算,每个距离计算包括了1024个维度浮点运算,总计要执行9000次,此外还得为测试向量准备2MB的存储空间。是否存在一种算法减少存储空间和计算时间的开销呢?k决策树就是k-近邻算法的优化版,可以节省大量计算开销。
本章小结 k-近邻算法是分类数据最简单最有效的算法,本章通过两个例子蒋旭了如何使用k-近邻算法构造分类器。k-近邻算法是基于实例的学习,使用算法时我们必须有接近实际数据的训练样本数据。k-近邻算法必须保存全部数据集,如果训练数据集很大,必须使用大量的存储空间。此外由于必须对数据集中的每个数据计算距离值,实际使用时可能非常耗时。
k-近邻算法的另一个缺陷是它无法给出任何数据的基础结构信息,因此我们也无法知晓平均实例样本和典型实例样本具有什么特征。可以通过概率测量方法处理分类问题以解决此问题。
参考资料:《机器学习实战》