一种流体动力的scRNA-seq条码技术,用于高通量的循环肿瘤细胞分析。
Introduction
循环肿瘤细胞(circulating tumor cells, CTCs)是一种罕见的转移癌细胞,能从原发性肿瘤上脱落进入循环系统,作为在远处组织形成继发性肿瘤的种子。CTCs被观察到极强的异质性在癌症细胞的上皮/间质转移过程中(另一篇与EMT相关的文章)。然而,每10 mL的血中有十亿计的红细胞和百万计的白血病却只有少于100个的CTCs。如何富集CTCs以下游分析是一个巨大的挑战。
免疫亲和性的富集方法通过CTCs的表面膜标记用于计数和下游分析,但是这种基于分子标记的方法使采样群体偏离,因此限制了综合分析的分子异质性。物理性质的富集方法如基于尺寸的选择,可以把CTCs从较小的红细胞和白细胞中分离而无需供选择的特定分子标记。然而,富集的细胞悬液通常依旧含有大量背景细胞(红细胞和白细胞),使其难以下游分析。尽管荧光染色结合免疫组化(IHC)、原位杂交(ISH)以及单细胞WB可能被用于研究CTCs的基因和蛋白表达,荧光成像的带宽限制了分子标记物的数目。
为了充分表征转移的CTCs的全转录组分析和通路调节,单细胞RNA测序(scRNA-seq)被用于毛细抽吸和介电泳微流控挑选的CTCs,其细胞被荧光染色标记。然而,这些技术依然被受限于它们有限的通量和基于抗体标记表达的依赖。常用的标记如上皮细胞粘附分子(EpCAM)和细胞角蛋白(cytokeratins)可能会错过大量不表达这些蛋白的CTCs。因此需要研究一种高通量的、无污染的scRNA-seq,以研究CTCs的通路调节和异质性。
为此,作者提出了Hydro-Seq,使用了基于尺寸的单细胞捕获,避免可能被分子CTC选择造成的偏差。并在21例晚期乳腺癌病人中验证,测序了666个CTCs,确定了肿瘤转移和治疗的关键生物标记的细胞异质性。
Fig. 1 Hydro-Seq, a high capture efficiency scRNA-seq platform for contaminated rare samples
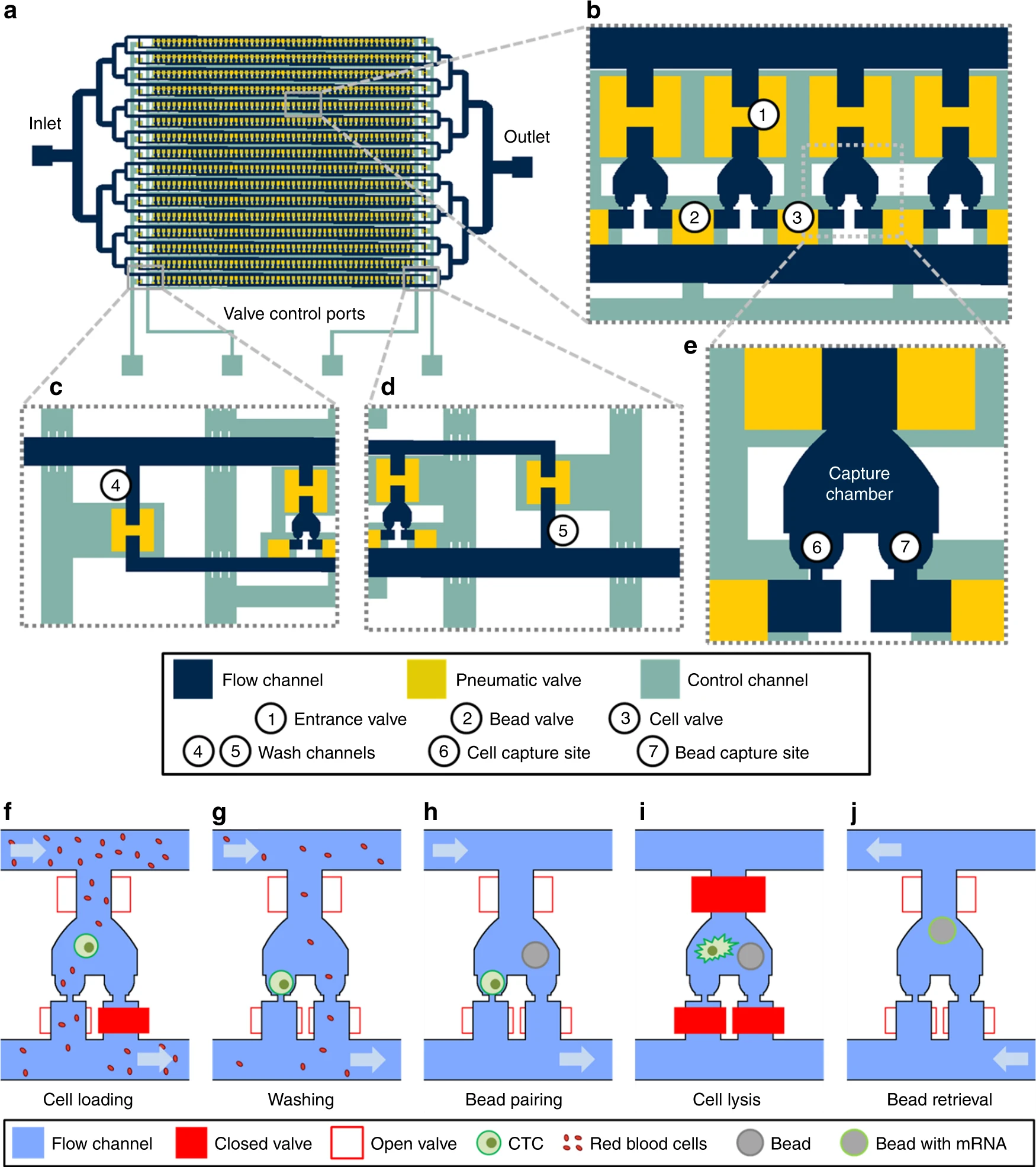
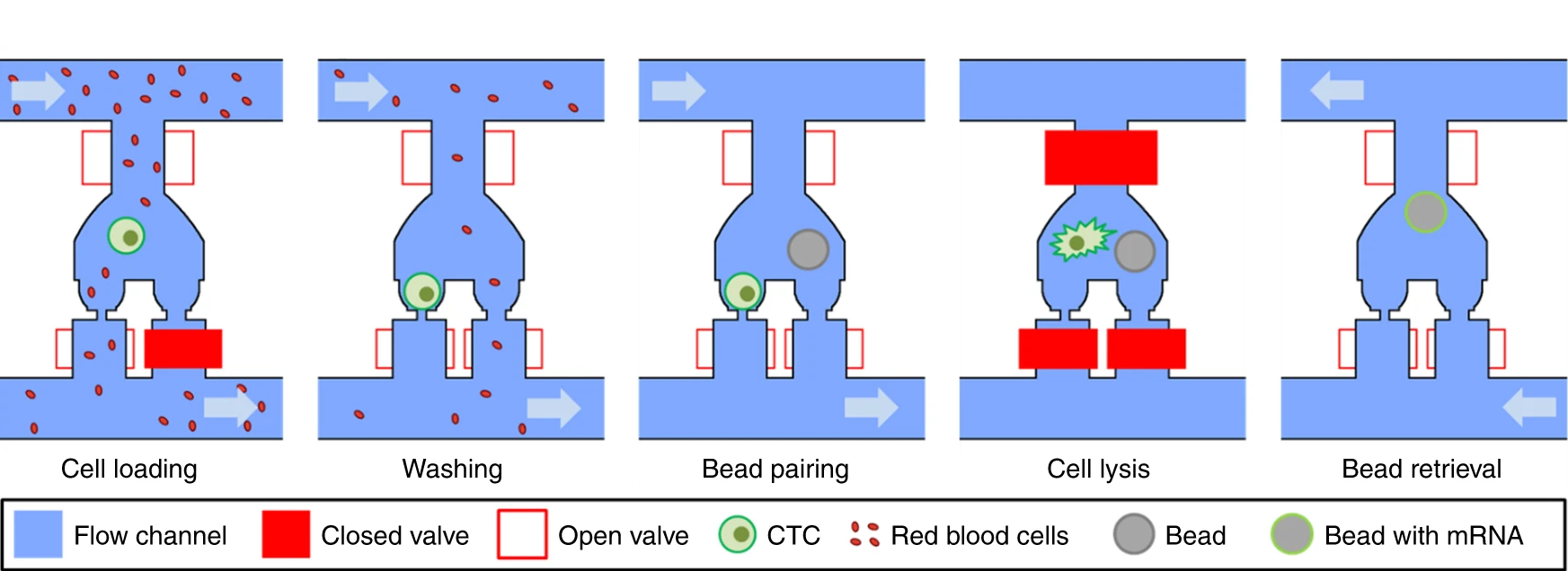
- (a)为了CTCs应用,Hydro-Seq被设计为16个平行、相同的分支通道。每个分支通道包含50个腔室用于小珠-细胞配对,每个芯片有800个腔室。
- (b)较近的视图突出了平行腔室的布置。为了最小化面积消耗,每个腔室和相邻的腔室分享阀门。入口阀峰高45 μm,面积200 × 200 μm能使小珠和细胞装载。细胞和小珠阀高15 μm,面积100 × 100 μm以用于高密度的腔室布置。
- (c, d)在分支通道的上下游,阀门控制的冲洗通道可以在样本装载和从进入通道传递裂解缓冲液时冲洗通道。
- (e)微流控的小珠-细胞配对腔室的放大视图。
- (f)小珠捕获阀在样本装载时关闭。较小的红细胞穿过捕获点,而较大的细胞(大于12 μm)堵塞了通道而被捕获。
- (g)细胞装载后,小珠捕获阀打开,通入PBS,将污染细胞洗出。
- (h)移除腔室的污染细胞后,装载小珠以配对。
- (i)裂解缓冲液被引入腔室,在关闭所有阀门后,腔室内产生紊流/湍流,小珠被移到细胞捕获区进行RNA捕获。
- (j)通过打开所有阀门,引入回流,小珠被回收至入口进行下游准备。
双端测序:科普。
Fig. 2 Species-mixing experiment and CTC loading in Hydro-Seq
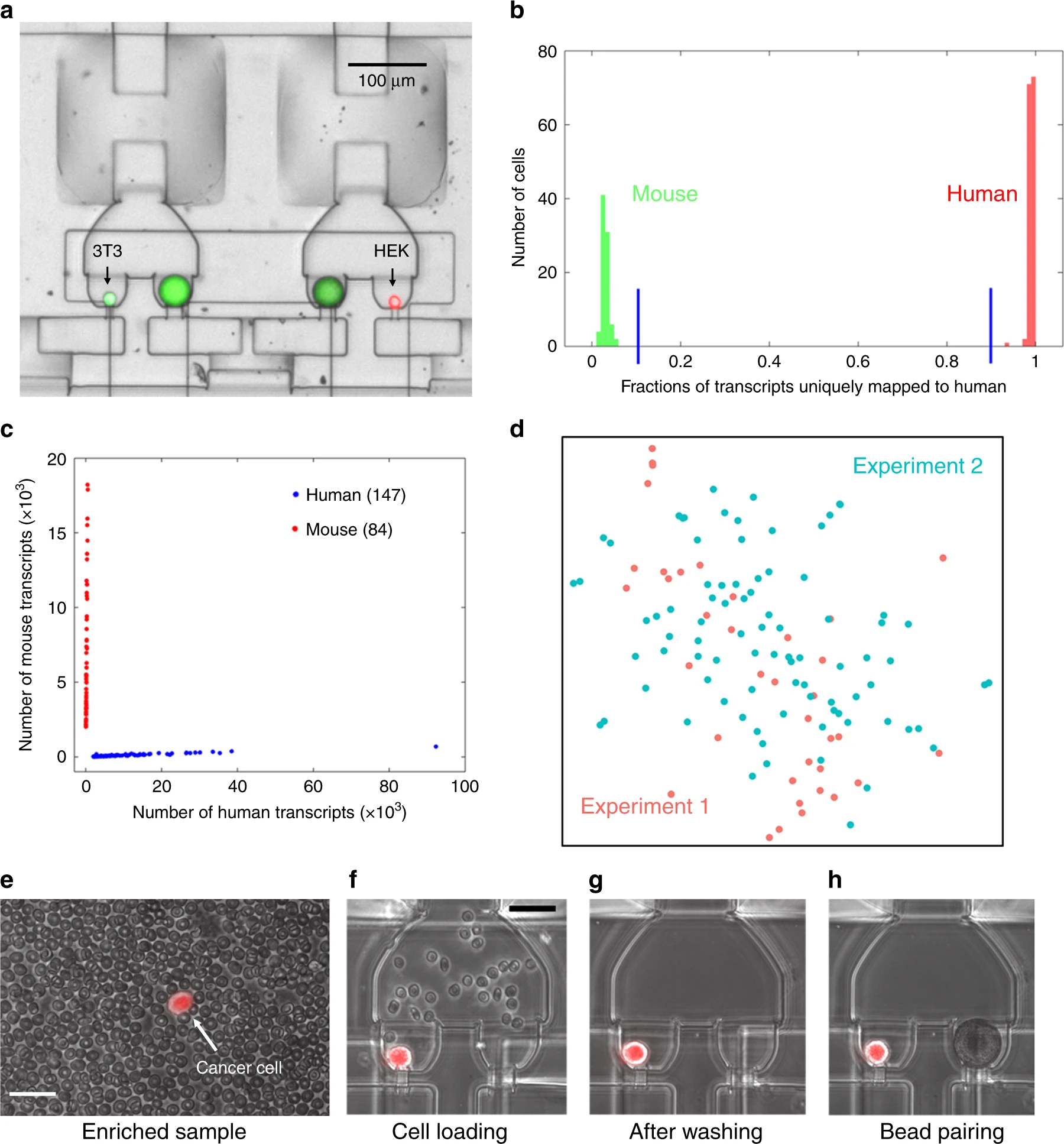
- (a)小珠与小鼠3T3细胞(绿色荧光)和人HEK细胞(红色荧光)配对的腔室图。荧光图像用于检查配对环境和确认捕获。
- (b)直方图用于展示混合的小鼠和人细胞的交叉物种污染。可以看到,超过90%的转录本被映射到人,标记为红色。超过90%的转录本映射到小鼠,标记为绿色。证明了该方法可以产生高生物特异性库。
- (c)混合物种分析强调了零细胞交叉污染的单细胞分辨率RNA测序。每个红点代表一个小鼠细胞,每个蓝点代表一个人类细胞。
- (d)2次独立的Hydro-Seq实验的HEK细胞的单细胞表达的tSNE图(实验一标记为红色,实验二标记为蓝绿色)。每个点代表一个细胞,2次实验的细胞均衡的分散,没有显示出任何簇,表明Hydro-Seq较好的复现性。
- (e)CTC富集(经过商业/之前的方法)后,CTC的scRNA-seq依然极具挑战性,原因是很多背景血细胞的存在。
- (f)样本装载中,红细胞流过腔室。
- (g)冲洗循环中,小珠阀打开,通过小珠捕获流道把其他血细胞移除。获得了无污染的单个孤立CTC以用于小珠配对。
- (h)小珠和单CTC配对以用于scRNA-seq。
Fig. 3 Immunostaining and single-cell RNA-sequencing of CTCs
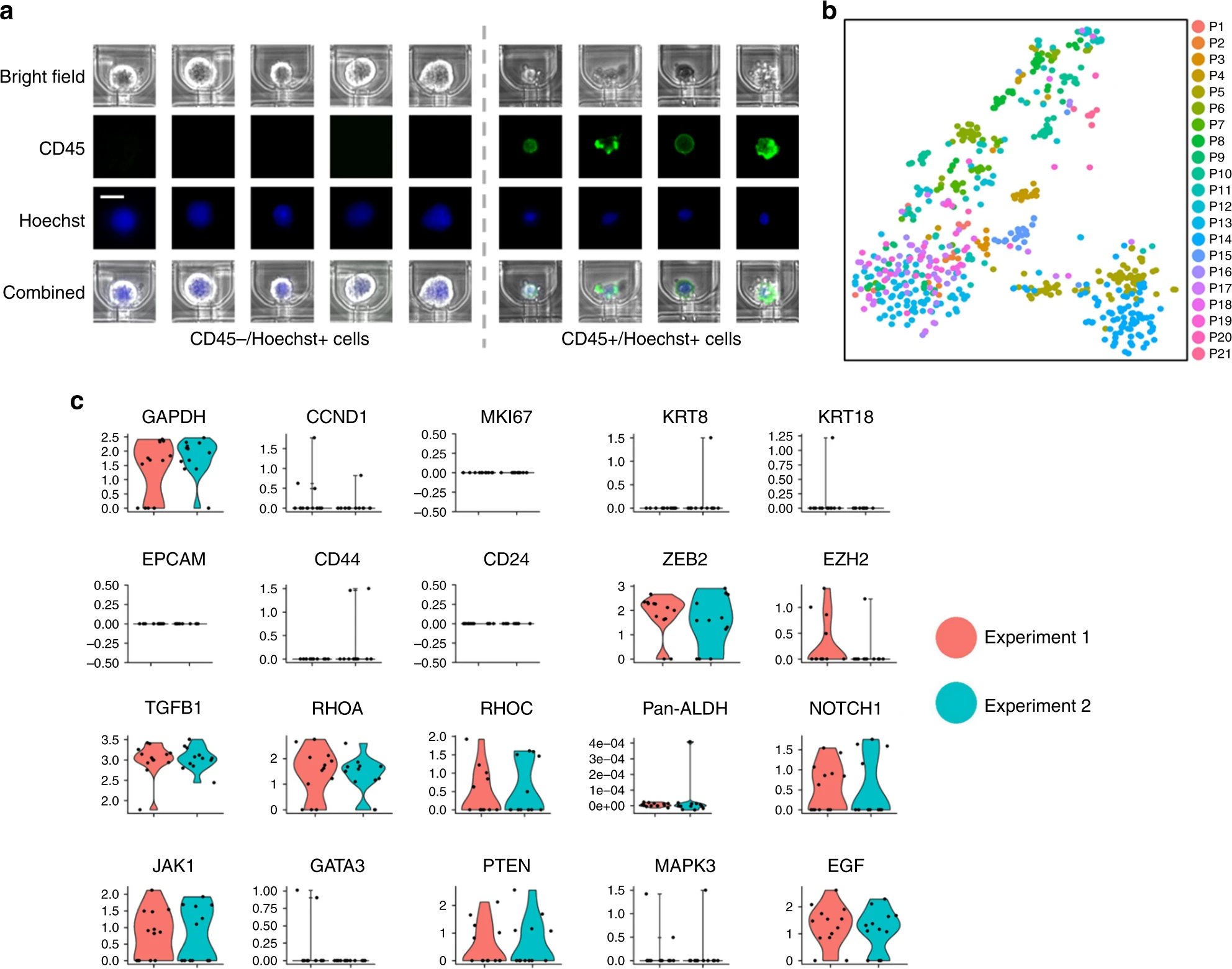
- (a)通过CD45和Hoechst染色,CD45阳性的成核细胞作为白细胞被识别。原因是一些较大的白细胞可能依然被捕获,因此使用Hoechst和CD45染色辨别,CD45+的成核细胞为白细胞,CD45-的成核细胞为假定的CTCs。
- (b)所有经Hydro-Seq处理的来自病人的CTCs样本的tSNE图(21个病人的666个CTCs,每个颜色代表一个独立病人样本)。
- (c)Hydro-Seq的复现性测试。同天处理同一个病人的两管采血液,两次实验取得了相当的CTC计数(实验一:13个CTCs,标记为红色;实验二:12个CTCs,标记为蓝绿色)。关于持家、细胞增殖、上皮、基质和其他基因的表达,显示了很好的复现性。
tSNE:T分布随机近邻嵌入,科普。
Fig. 4 Gene expression and clustering of breast CTCs
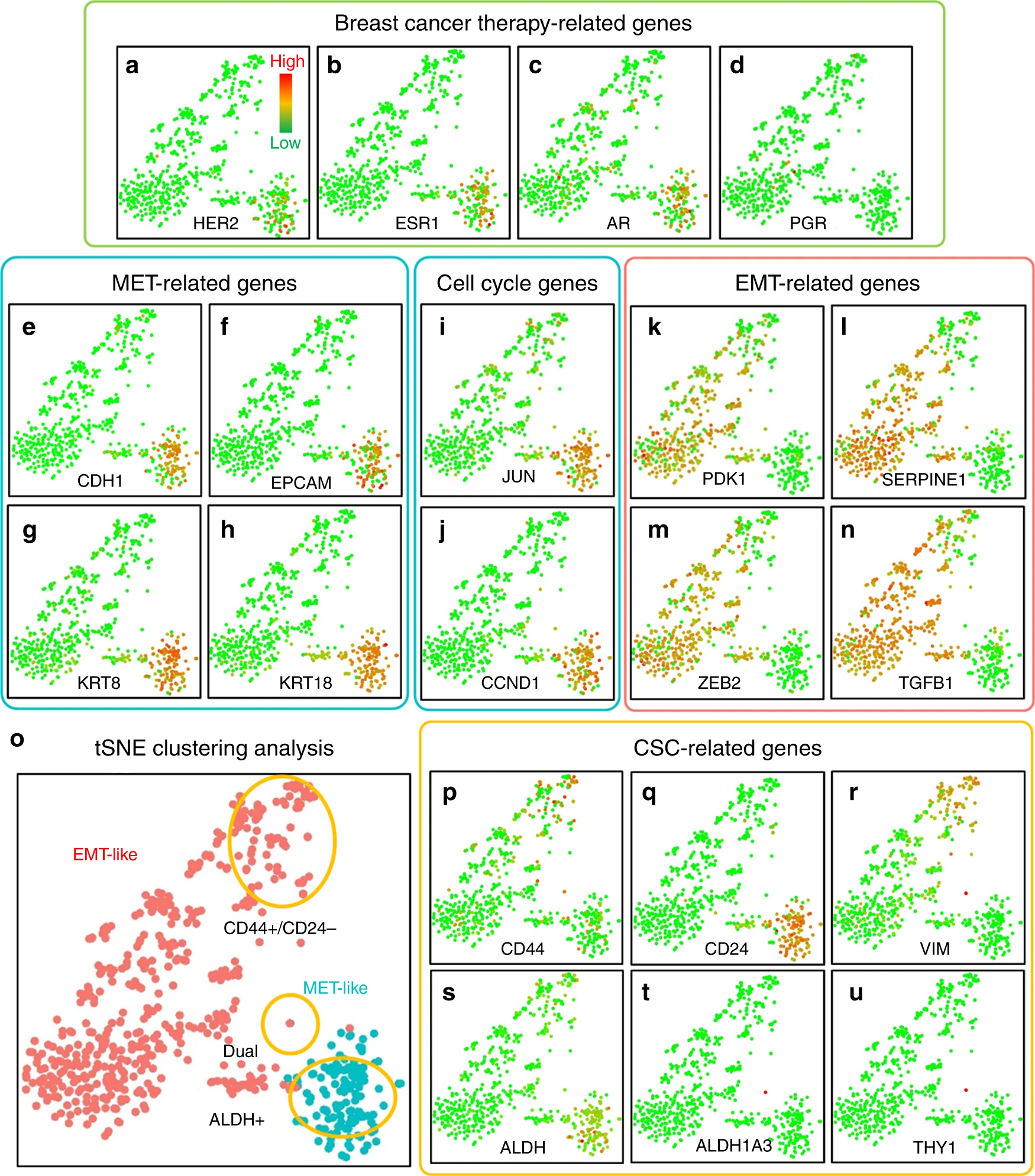
(a-d)乳腺癌治疗相关基因。
- (a)人表皮生长因子受体2(HER2/Erbb2)。
- (b)雌激素受体(ESR1)。
- (c)雄激素(AR)。
- (d)孕激素受体(PGR)。
(e-h)MET相关基因。
- (e)E-钙黏蛋白(CDH1)。
- (f)上皮细胞黏附分子(EPCAM)。
- (g)角蛋白8(KRT8)。
- (h)角蛋白18(KRT18)。
(i, j)细胞周期基因。
- (i)c-jun(JUN)。
- (j)细胞周期蛋白D1(CCND1)。
(k-n)EMT相关基因。
- (k)磷酸肌醇依赖性激酶1(PDK1)。
- (l)纤溶酶原激活物抑制剂-1(SERPINE1)。
- (m)EMT转录因子(ZEB2)。
- (n)转化生长因子-β(TGFB1)。
(o)HER2阳性、MET样和HER2阴性、EMT样CTCs的聚类和分离。
(p-u)干细胞特性相关基因。
- (p)CD44。
- (q)CD24。
- (r)波形蛋白(VIM)。
- (s)泛乙醛脱氢酶亚型(ALDH)。
- (t)ALDH1a3。
- (u)CD90 (THY1)。
每个点代表一个CTC。绿点代表最低表达,红点代表最高表达。表达做了对数正则化。
CD:cluster of differentiation,分化簇。
Fig. 5 CTC intra-patient heterogeneity of EMT-like and MET-like states
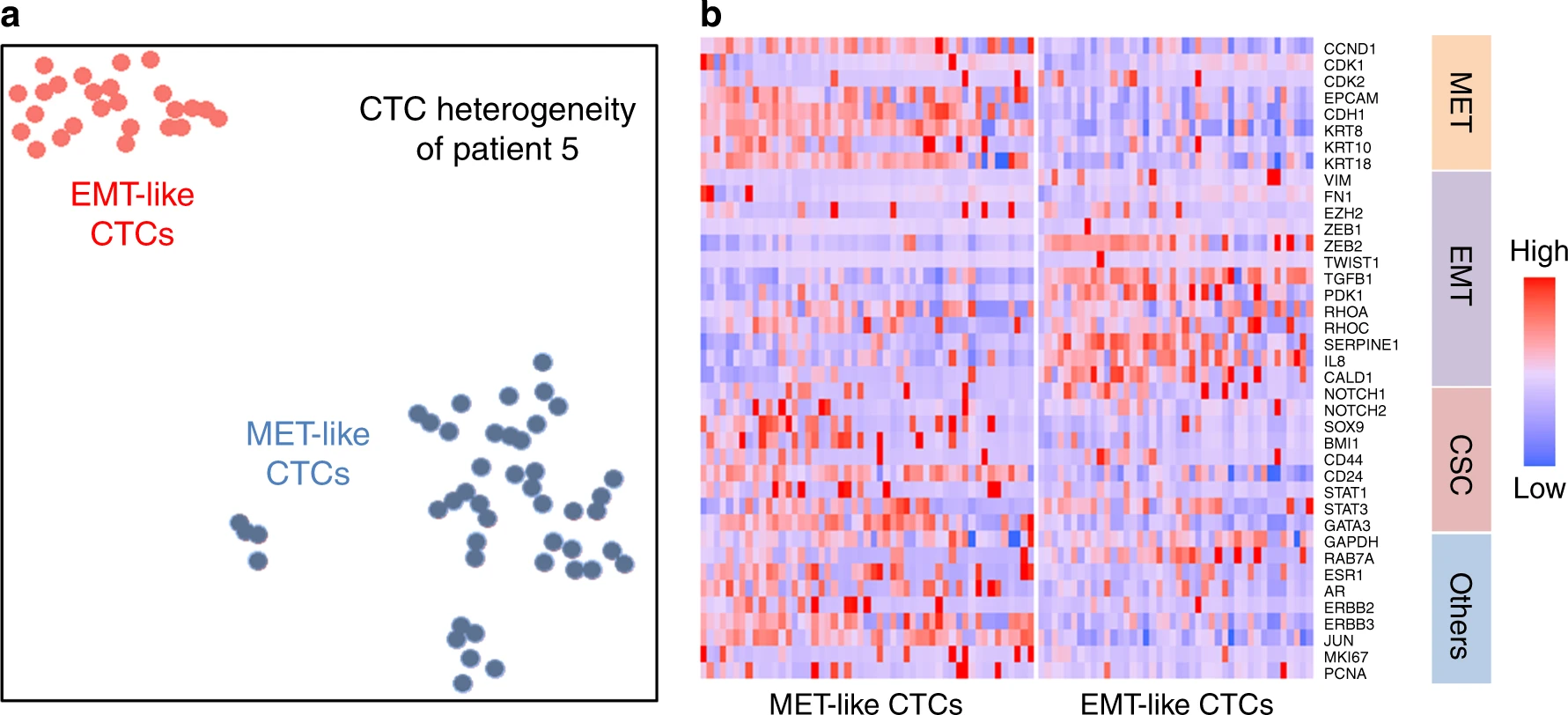
- (a)来自一个病人(5号)样本的78个CTCs经过tSNE聚类绘制如下,证明了MET-like和EMT-like的CTC群有清楚的分界。
- (b)热图显示了2簇细胞的显著基因表达(EMT、MET、CSC相关基因)差异。
Table 1 Pathways distinguishing the HER2+ MET-like and HER2− EMT-like CTCs. The p-value was determined by Fisher’s Exact Test
| Top-ranked pathways | p-value | Roles |
|---|---|---|
| Integrin-linked kinase signaling(整合素连接激酶信号) | 2.17E-08 | EMT/MET |
| Stabilization and expansion of the E-cadherin(上皮型钙黏蛋白) adherens junction | 3.66E-06 | EMT/MET |
| Signaling events mediated by focal adhesion kinase(粘着斑激酶) | 1.21E-04 | EMT/MET |
| Validated targets of C-MYC transcriptional activation | 1.38E-04 | CSC |
| PDGFR-beta(血小板衍生生长因子受体β) signaling pathway | 6.56E-04 | CSC |
| AP-1 transcription factor network | 5.88E-04 | |
| RhoA signaling pathway | 3.28E-04 | CSC |
| Regulation of Telomerase(端粒酶) | 1.73E-03 | CSC |
| ErbB1 downstream signaling | 2.22E-03 | CSC |
| PAR1-mediated thrombin signaling events | 1.16E-03 |
Discussion
传统的研究CTC基因和蛋白表达的方法为荧光成像结合原位杂交(ISH)和免疫组织化学(IHC)。由于荧光成像的带宽限制, 多重荧光分析一次只能达到6-7个通道(或标记)。考虑到CTC识别的3个标记,只要3-4个通道能用于研究。尽管可以移除抗体并应用新的以染色,整个流程重复且难以扩大。可替代的方法对CTCs的全转录组分析是使用一系列的细胞分选术。CTCs通常由上皮细胞黏附分子(EpCAM)或广谱细胞角蛋白(pan-cytokeratin)染色标记,单细胞操纵方法比如毛细管抽吸和介电泳微流控被用于分配靶细胞去不同的管以用于下游处理。尽管这种方法成功对CTCs进行scRNA-seq,然而一系列挑选方法费力、低通量而且难以扩大每个样本分析的数量。此外,EpCAM和pan-cytokeratin的分子选择可能丢失重要的CTC亚群如本文提出的间质CTCs。
因此,在使用传统方法研究CTC时,存在细胞和基因数目的权衡。荧光ISH和IHC实验达到高通量的CTCs分析但是限制了可供分析的生物标记数目,阻碍了对治疗发展的通路调节研究。scRNA-seq的细胞分选使CTCs的全转录组分析成为可能然而限制了分析的细胞数量以及对细胞异质性的研究,而这将是肿瘤细胞的关键表征。
为了克服这些限制,Hydro-Seq取得了CTCs的高通量和全转录组分析,以了解细胞异质性和阐述细胞调控途径。其具备的优点:1.对稀有细胞的高捕获率。2.移除污染细胞能力强。
Reference
Cheng Y-H, Chen Y-C, Lin E, et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells[J]. Nature Communications, 2019, 10(1): 2163.