一种整合了捕获,分离和分析细胞膜标记物以及通过单CTCs和CTC簇定量蛋白质分泌的微流控方法。
Introduction
上皮癌的转移级联反应是一个复杂的生物学过程,在不同类型的癌症之间具有巨大的动力学和运动的多样性。近年来关注重点已经转移到对循环肿瘤细胞(CTCs)的分析。
对于个性化癌症治疗,详细了解每个癌症病人的特征是非常重要的。从癌细胞上脱落的CTCs、自由循环DNA和细胞外囊泡是关于肿瘤的有价值消息,因此被提议为诊断测试的靶点。目前,主要使用先进测序工具的分析技术,通过DNA和RNA分析中推断潜在的药物敏感性。然而,最近的研究显示了这些测量的结果与疾病结果或药物反应的低相关性。因此,CTCs的直接蛋白分析可能是更详细了解与药物敏感性相关的生物学过程的重要财富。
当前大多数定量CTCs细胞内和分泌蛋白的方法需要人工转移分离的CTCs去第二个分析仪器。不仅需要人工操作,也在分析前后引入了测量误差或者损害CTCs。除先前提到的技术障碍外,尚不清楚分离的CTCs是否能在常规细胞培养环境(即培养基和氧气浓度)的短期培养下存活下来。尽管有些研究报道效率超过50%,迄今为止CTC的培养成功率普遍很低,只有少数CTC来源的细胞系被建立。一个允许分离CTCs和后续分析而不需要进一步转运或处理的方法将克服上述瓶颈。
这里提出了一种整合了尺寸选择捕获和功能性分析CTCs的单个微流控平台。我们通过确定HER-2,EpCAM和白细胞(WBC)标记CD45的存在来明确区分CTCs和其他血液成分。此外,我们利用了基于磁珠的免疫分析,为定量分析少数甚至是单细胞的蛋白分泌开辟了道路。
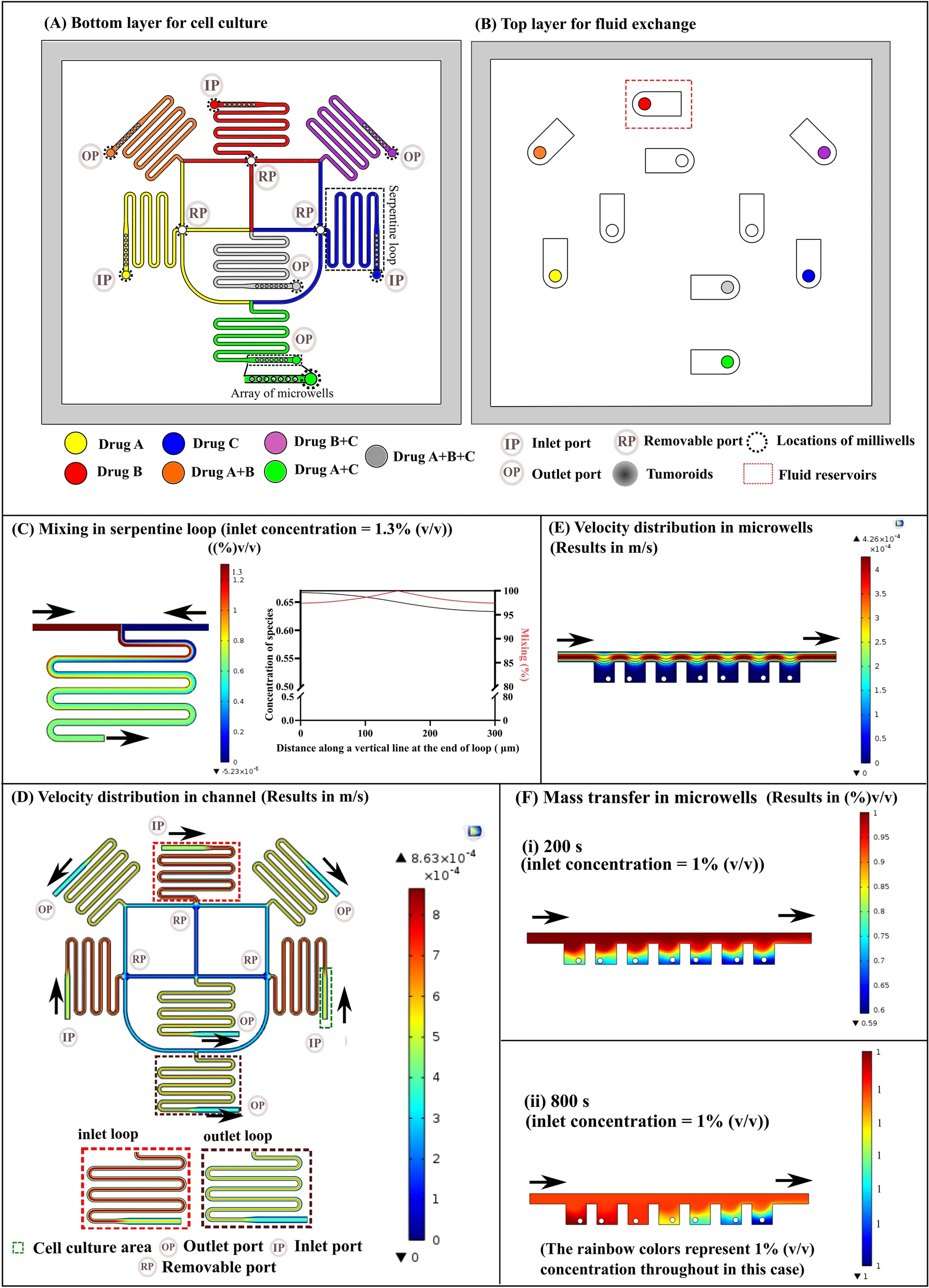
Figure 1. Microfluidic chip design and operation for CTC capture and analysis
- (a)测量体系包含微流控芯片、可以安装在显微镜载物台上的芯片架和带有永磁铁的盖子组成。照片还显示了分别建立与注射泵和压力系统的流体和气动连接的管道。
- (b)30个分析室的子集的显微照片。每个芯片总共容纳1152个CTC分析室,该室分为四部分,每部分有3排96列。
- (c)一个分析室的中心放大,在这里可以实现CTCs的尺寸选择性捕获和磁珠捕获。图中,一个CTC‐WBC簇和单个磁珠被捕获并染色。这两条可见的浅灰色线是帮助CTC捕获细胞陷阱的微柱。
- (d-h)工作流程示意图;虚线表示微柱的位置。
- (d)首先,全血中的CTCs被捕获在腔室中间的流体收缩处。
- (e)连续用DMEM洗涤。提供磁珠并与CTCs共固定化。
- (f)带磁铁的盖子被放置在微芯片的顶部,以吸引磁珠。
- (g)接下来,驱动阀门形成容积为80 pL的分析室。在孵育期间,分泌的细胞因子G‐CSF与磁珠表面的抗G‐CSF抗体结合。最后,清洗腔室,进行标签化。
- (h)经过最终清洗后,微流控芯片可以成像。在此期间不需要磁铁。
- (i)用于检测G-CSF的三明治免疫法的示意图。
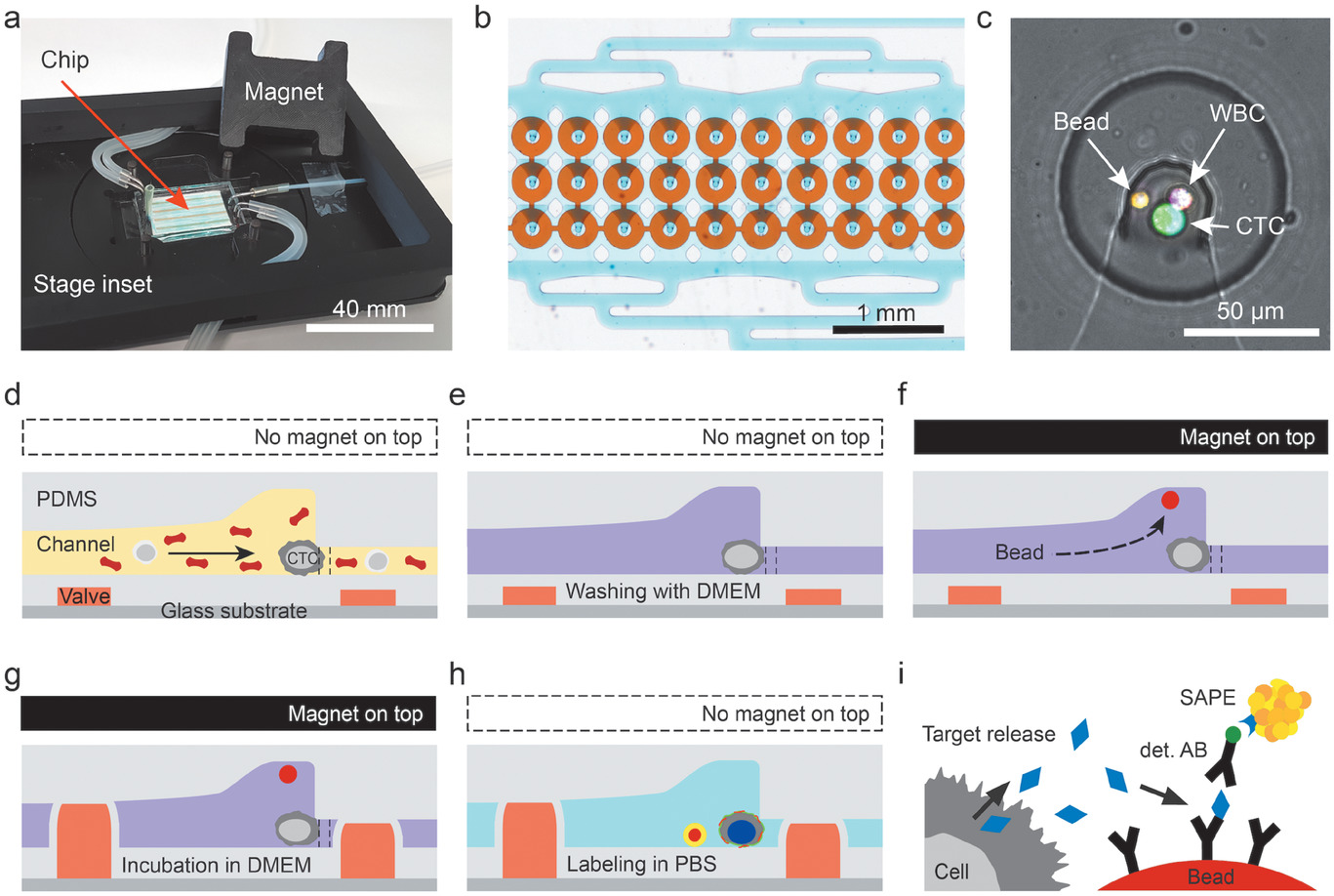
Figure 2. Determination of the CTC capture efficiency
- (a)展示了首先在出口通过记录荧光显微镜图像然后随后对分析室拍照并分析以计算捕获细胞,检测未捕获细胞的工作流程。这些测量中,细胞被calcein AM染色或表达胞质GFP。
- (b)通过分析出口记录的图像获得的荧光轨迹图。
- (c)捕获效率和流速的关系。
- (d)限制流速为50 μL min-1时,捕获效率和通道陷阱高度的关系。
- (e)优化完毕后(陷阱高度:7.5 μm,流速:20 μL min-1)时,捕获效率和各种细胞系的关系。
- (f)单个MCF-7细胞和不同大小细胞簇的个体捕获效率。
- (g)通过施加反向1000 μL min-1流速的含1%BSA的PBS来释放CTC的效率。但20倍于血液处理的流速对细胞施加了高剪切力和压力。
以上结果显示:所有模型细胞的捕获效率均在95%以上,但总体释放率仅为67%。因此对捕获细胞进行芯片上的分析,比将其释放到下游分析更科学。
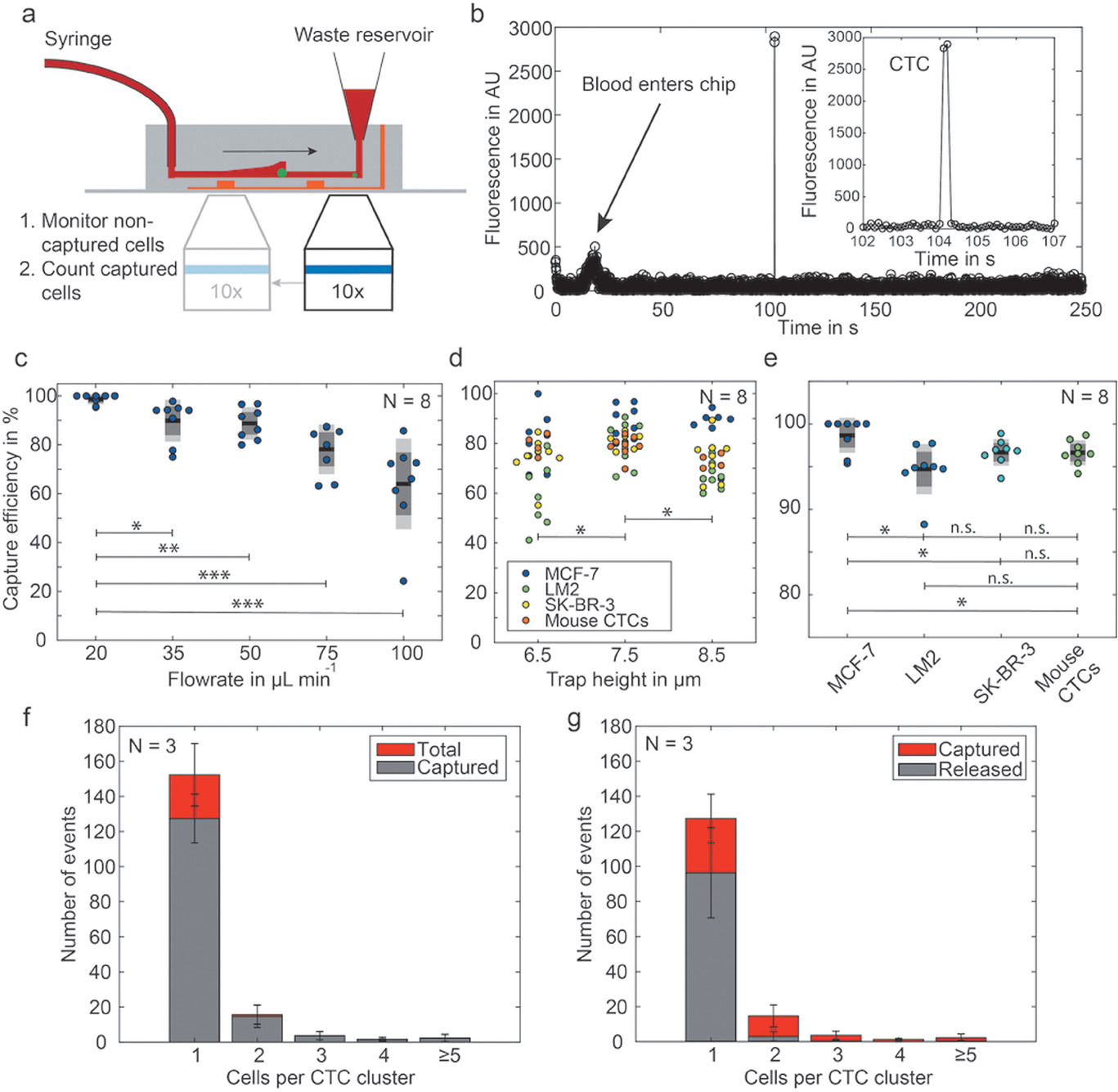
Figure 3. Optimization of the G-CSF assay on fluorescently barcoded magnetic beads
- (a)三明治夹心免疫法的示意图,这是在被捕获抗体功能化的磁珠上进行的。生物素化的检测抗体(det. AB)被荧光标签标记。最终选定为链霉亲和素-藻红蛋白(SAPE)。
- (b)被捕获到一个单独的微腔的磁珠荧光信号。使用条码珠去明确识别和定位磁珠。
- (c)G-CSF在微孔板上进行三明治夹心免疫法。使用了赛默飞公司(TF)的两种不同的试验:标准试验和高敏感性(HS)试验。绘制了校准曲线。
- (d)比较了两种供应商的检测抗体,分别用TF的SAPE或荧光球标记。
- (e)用于定量CTCs的G-CSF产量的最终校准曲线,最低检测限为1.5 ng mL-1。
Figure 4. Brightfield (first column) and fluorescence images of the trapping site, occupied by individual cells of the investigated cell lines, and for comparison, a trapped WBC (bottom row)

捕获位点的明场(第一列)和荧光图像,被研究的细胞系的单个细胞占据,以及作为比较的一个捕获的白细胞(最底部)。伪彩色的荧光图揭露了有核细胞的存在(NucBlue)和膜蛋白HER-2、CD45、EpCAM的存在和缺失以及磁珠捕获的G-CSF的分泌。磁珠被荧光比率识别在658 nm(条码 1)/712 nm(条码2)。
Figure 5. Protein expression of single cells and mouse model CTCs
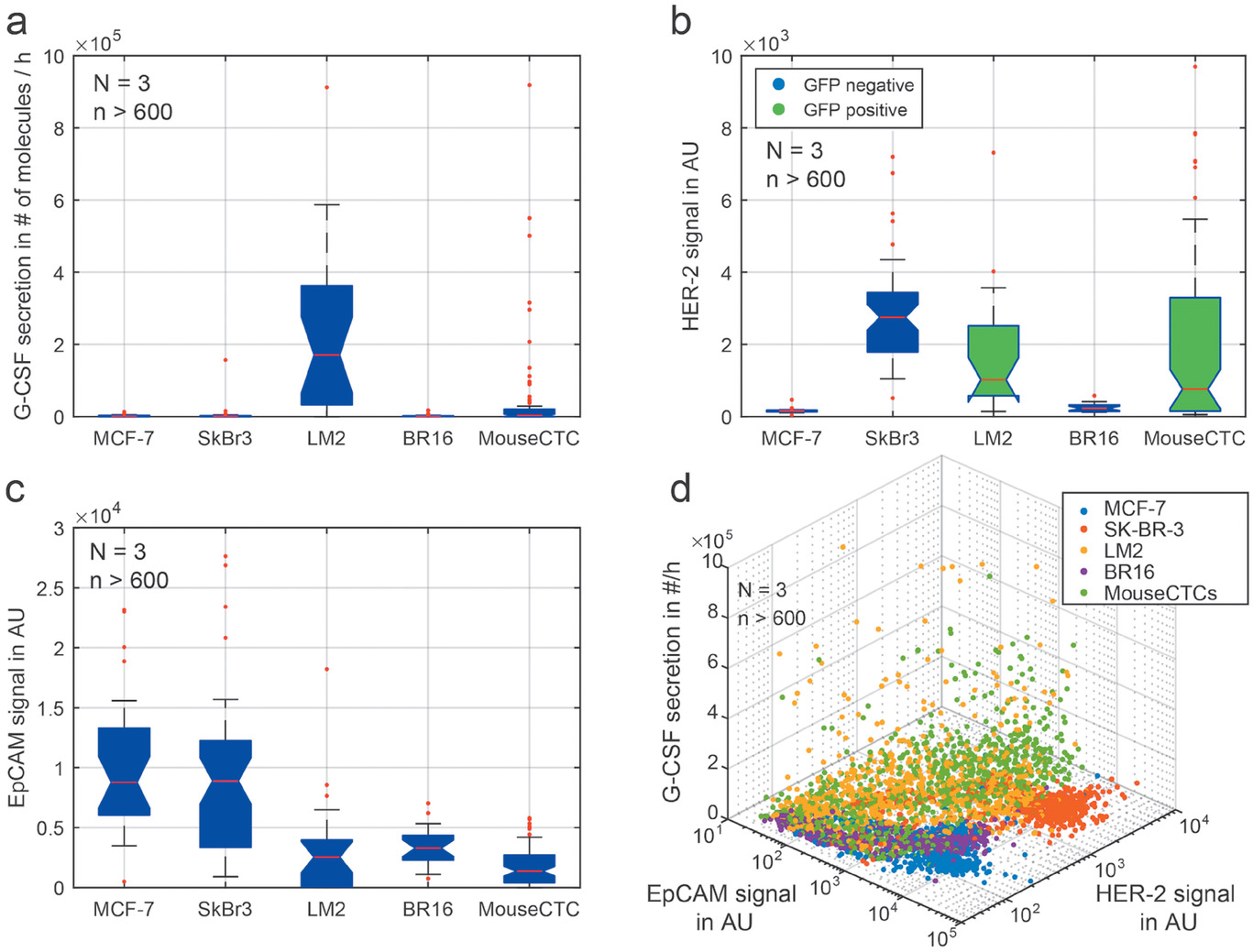
- (a)4小时孵育后定量分析G-CSF分泌水平。MCF-7、SK-BR-3和BR16细胞中没有检测到G-CSF蛋白,而LM2细胞存在多种G-CSF高表达的类型。
- (b)5种测试细胞系的HER-2表达比较。LM2细胞系和鼠模型CTCs表达GFP,模糊了HER-2表达。HER-2在SK-BR-3细胞中表达最高,而在MCF-7和BR16细胞则较低。
- (c)所有测试细胞系的EpCAM表达比较。EpCAM的表达在MCF-7细胞中含量最高,其次是SK-BR-3和BR16细胞,而其他细胞系较低。
- (d)将所有标志物关联起来,可以实现不同细胞系的区分。
Conclusion
作者开发了一个能够从全血中进行CTC分离且组合定量分析G-CSF分泌的微流控芯片。与其他方法相比,该平台通过免疫组化和磁珠上ELISA可以同时进行多个膜蛋白和分泌因子的高灵敏分析。为后续直接进行单细胞蛋白质组分析研究提供了可能,有助于更深层次的理解癌症转移的生物学机理。
Reference
Armbrecht L, Rutschmann O, Szczerba B M, et al. Quantification of Protein Secretion from Circulating Tumor Cells in Microfluidic Chambers[J]. Advanced Science (Weinheim, Baden-Wurttemberg, Germany), 2020, 7(11): 1903237.