一种简单的、使用带有气动微结构的集成微流控装置,以高通量、几何动态和多图案方式高度可控地固定蛋白质和细胞。
Introduction
微图案技术在生物化学和细胞分子生物学/免疫学领域的应用一直是人们关注的热点。选择性的排列生物物质在指定的基质表面获得了大量基础细胞研究的关注,也开启了在生物医学诊断、药物发现和组织工程实验方法的大门。例如,最近在细胞和生物分子微图案方面的进展已经证明,在增加我们对同型/异型生物群落的结构和功能关系的理解方面是非常宝贵的。
在过去的十年中,已经探索和提出了多种微图案策略,如光刻、模板图案、微流控图案、微接触印刷、喷墨印刷和选择性等离子体蚀刻,在高通量和几何可控的方式用于在微控制系统的发展和创新。尽管如此,对于动态目的的多因素组织和微图案的系列化操纵仍在探索之中,这就需要对生物微环境的构建进行系统的考虑。
整合微图案和微流控方法并不容易。微图案表面通常与微流控系统不兼容,因为其严格的处理(热或者等离子体结合)可能会改变微图案表面的性质和功能,或者甚至损坏蛋白质和细胞的生物活性。因此,在微流控装置内发展原位微图案可能是有益的。另一方面,微流控系统不断增加的复杂性也要求开发功能表面的稳健制造和控制方法。目前,已有几种方法如层流微图案成功地控制了微通道的表面性质和细胞粘附。此外,利用压力调节的微流控阀已经开发出了原位微流控蛋白质捕获技术。然而,无论是空间控制(如几何形状和大小可变)和高通量,还是多种生物组分定位(即不同类型的蛋白质和细胞)的微流控中微图案化在很大程度上仍然无法实现。
为此,作者提出了一种通过气动微结构(PμSs)和PμS辅助表面钝化,用于蛋白质和细胞的原位微图案化。根据PμSs的可调驱动作用,可将不同数量的定位蛋白和细胞排列在微室中。此外,利用特殊设计的PμS阵列可实现多种生物组分的高通量、几何可控地微图案化。此外还证明了这种方法能够通过PμSs的连续操作,在同一腔室中同时完成不同蛋白质/细胞的有组织定位地共同微图案化。
Fig. 1 Micropatterning of proteinsand cells in the integrated microfluidic device by using pneumatic microstructure (PμS) actuation and spatially controlled surface passivation

- (A)PμSs随气压开关的三维视图。
- (B)装置的组成(从上到下依次显示四层,即流体层,控制层,支撑层和载玻片)。
- (C)通过Pluronic F127进行PμS辅助的表面钝化,该过程仅修改了腔室中的非阻塞区域。
- (D)在PμS阻塞的表面上进行蛋白质微图案化。荧光图像显示FITC标记的BSA固定在与PμS阻塞区域相对应的圆形表面上。
- (E)在PμS阻塞和粘附蛋白预处理过的表面上进行细胞微图案化。 HUVEC-C细胞在与PμS阻塞区域相对应的表面上表现出良好的粘附控制。右上角正方形中的示意图显示了具有亲水和疏水基团的Pluronic F127的功能,并显示了其固定在PDMS表面上的情况。
有关亲疏水性对细胞粘附影响:见此。
Fig. 2 Dynamic investigation of PμS manipulation and micropatterning of proteins and cells at different gas pressures
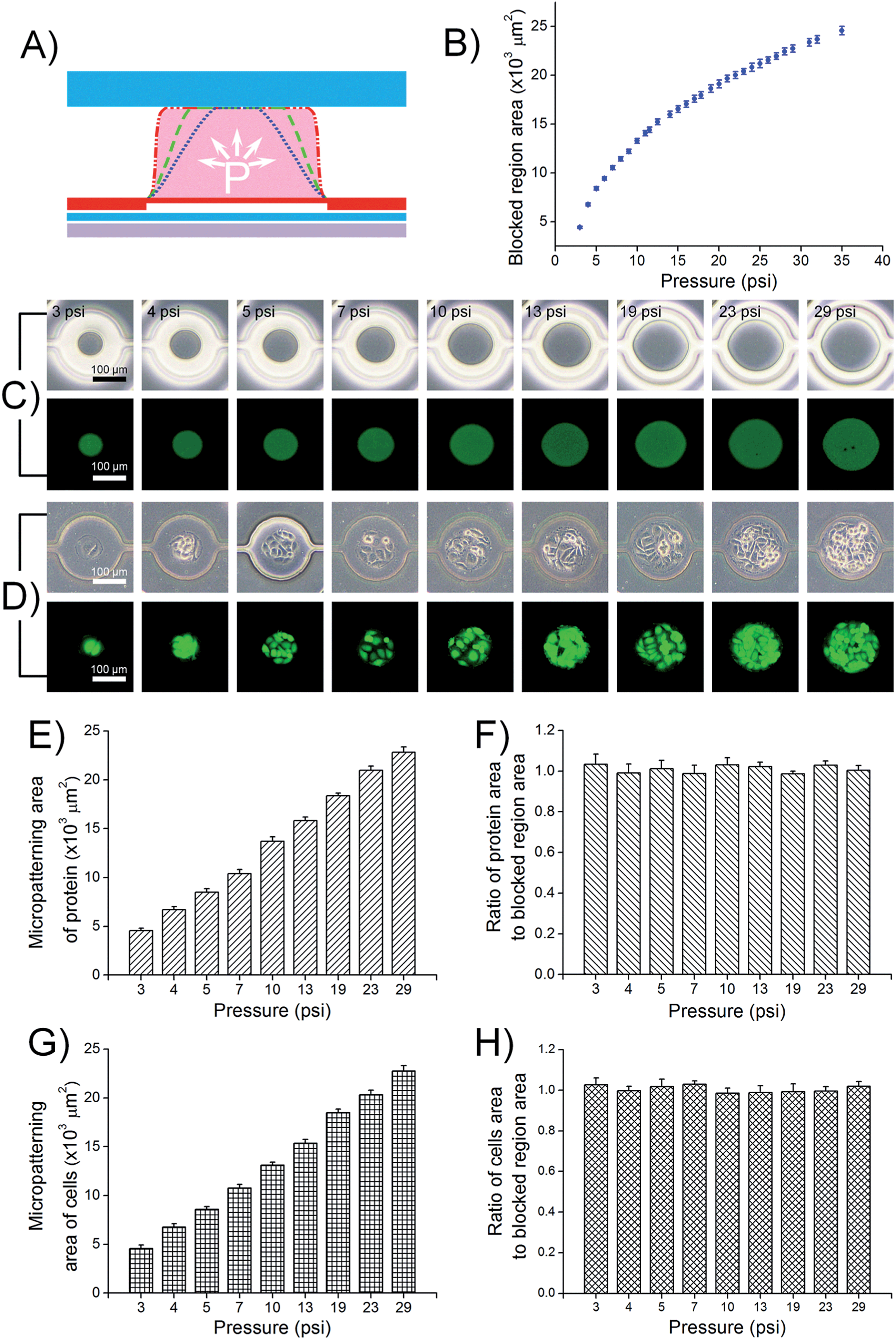
- (A)气体压力变化下PμS变形动力学的剖视图。
- (B)定量分析各种气体压力下的PμS阻塞动力学。将PμS和微腔顶面之间的阻塞区域用于定量评估。
- (C)在各种气压下,PμS阻塞区域的光学图像(顶部)和FITC标记的BSA微图案的荧光图像(底部)。
- (D)HUVEC-C细胞微图案的光学图像(顶部)和FDA/PI溶液染色的HUVEC-C细胞微图案的荧光图(底部)。
- (E)BSA在各种气压下的微图案面积。
- (F)BSA图案区域与PμS阻塞区域之间的比率。
- (G)HUVEC-C细胞在各种气压下的微图案面积。
- (H)HUVEC-C细胞图案区域与PμS阻塞区域之间的比率。
Fig. 3 High throughput micropatterning using the PμS array
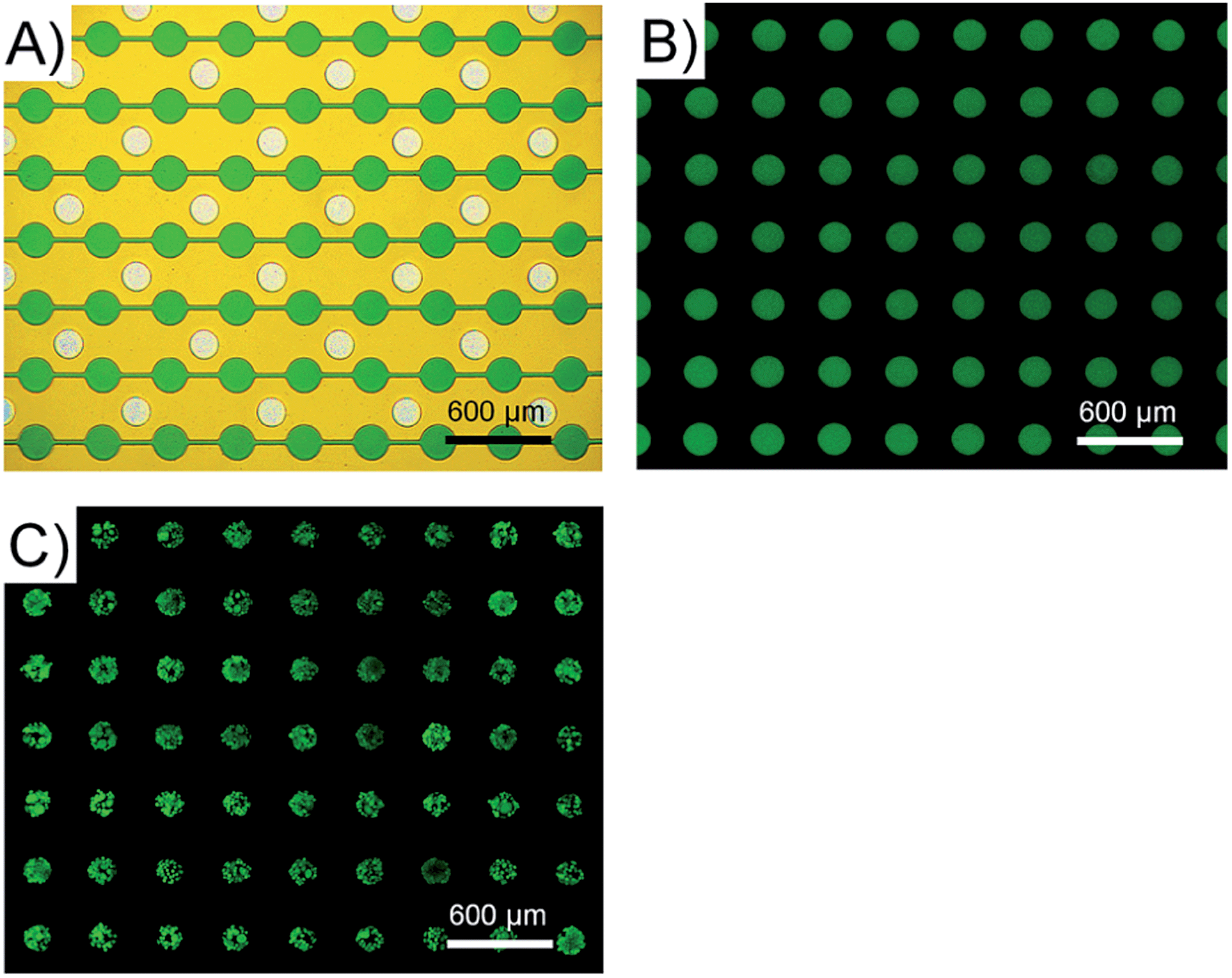
- (A)PμS阵列的光学图像。在控制层的微通道网络中加载了绿色食用染料,用于PμSs的可视化。
- (B)腔室中FITC标记的BSA的阵列状微图案。
- (C)腔室中HepG2细胞的阵列状微图案化(AO/PI染色)。这些结果表明,可以通过使用PμS阵列以高通量的方式定位蛋白质和细胞。
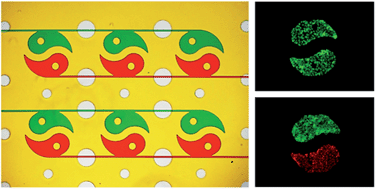
Fig. 4 The shape-diversified PμSs in the microfluidic devices

微流控装置中形状各异的PμSs。本研究共制备了6种PμSs,如圆形、条形、椭圆形、波浪形、方形和太极形PμSs。两组装置中的PμSs分别用绿色和红色食品染料进行标记。
Fig. 5 Geometry-controlled micropatterning by different well-designed PμSs
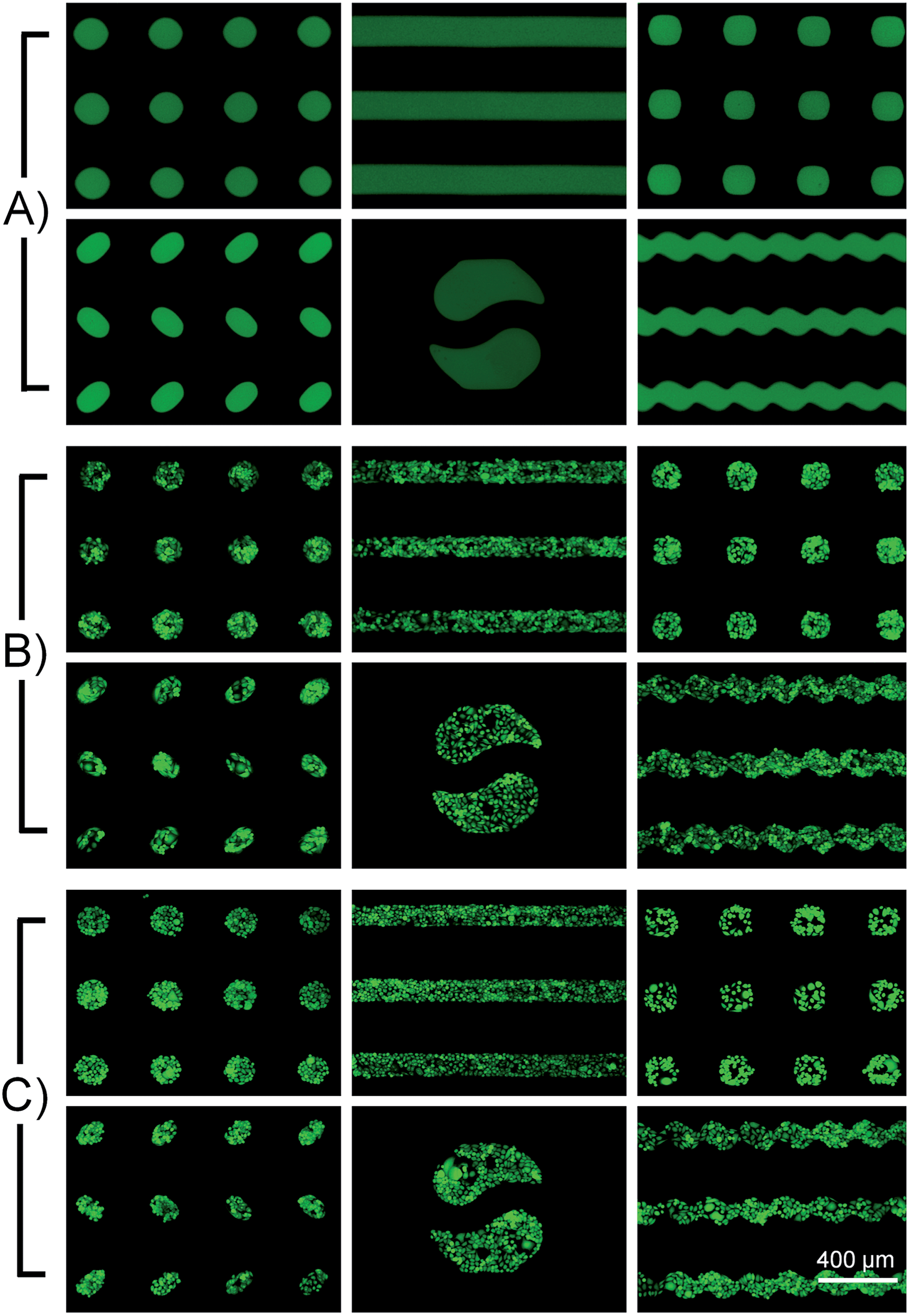
- (A)使用不同的PμSs进行BSA微图案化的荧光图像。
- (B)使用不同的PμSs进行HUVEC-C细胞微图案化的荧光图像。
- (C)使用不同的PμSs进行HepG2细胞微图案化的荧光图像。
Fig. 6 Co-micropatterning of different proteins/cells using PμSs

- (A)将具有不同形状的FITC标记的BSA和Alexa Fluro594标记的CEA固定在同一腔室中。
- (B)在同一腔室内以各种形状定位HUVEC-C细胞(红色)和HepG2细胞(绿色)。
Conclusion
在这项研究中,作者提出了一个原位微图案化方法,通过使用气动开关和可控表面钝化在微流控装置中实现蛋白质和哺乳动物细胞的高通量和多重定位。
- 气动微结构可以基于精确控制的尺寸动力学,方便地在定义明确的区域中实现多种蛋白质/细胞固定化。
- PμSs可以完成装置中多个生物成分的阵列状微图案化操作。
- 精心设计的PμS具有简单实现蛋白质/细胞定位的几何多样性的能力。
- 证明了在微流控设备中微腔的定义表面上可以通过形状调节排列来共同图案化多种类型的蛋白质和细胞,这将有助于涉及蛋白质等多种相互作用的生物学研究,细胞共培养和交流和组织模拟。
- PμS辅助微图案的片上集成可能对开发各种微流控高通量筛选,临床诊断和免疫传感很有用。
Reference
Wang J-C, Liu W, Tu Q, et al. High throughput and multiplex localization of proteins and cells for in situ micropatterning using pneumatic microfluidics[J]. Analyst, 2015, 140(3): 827–836.