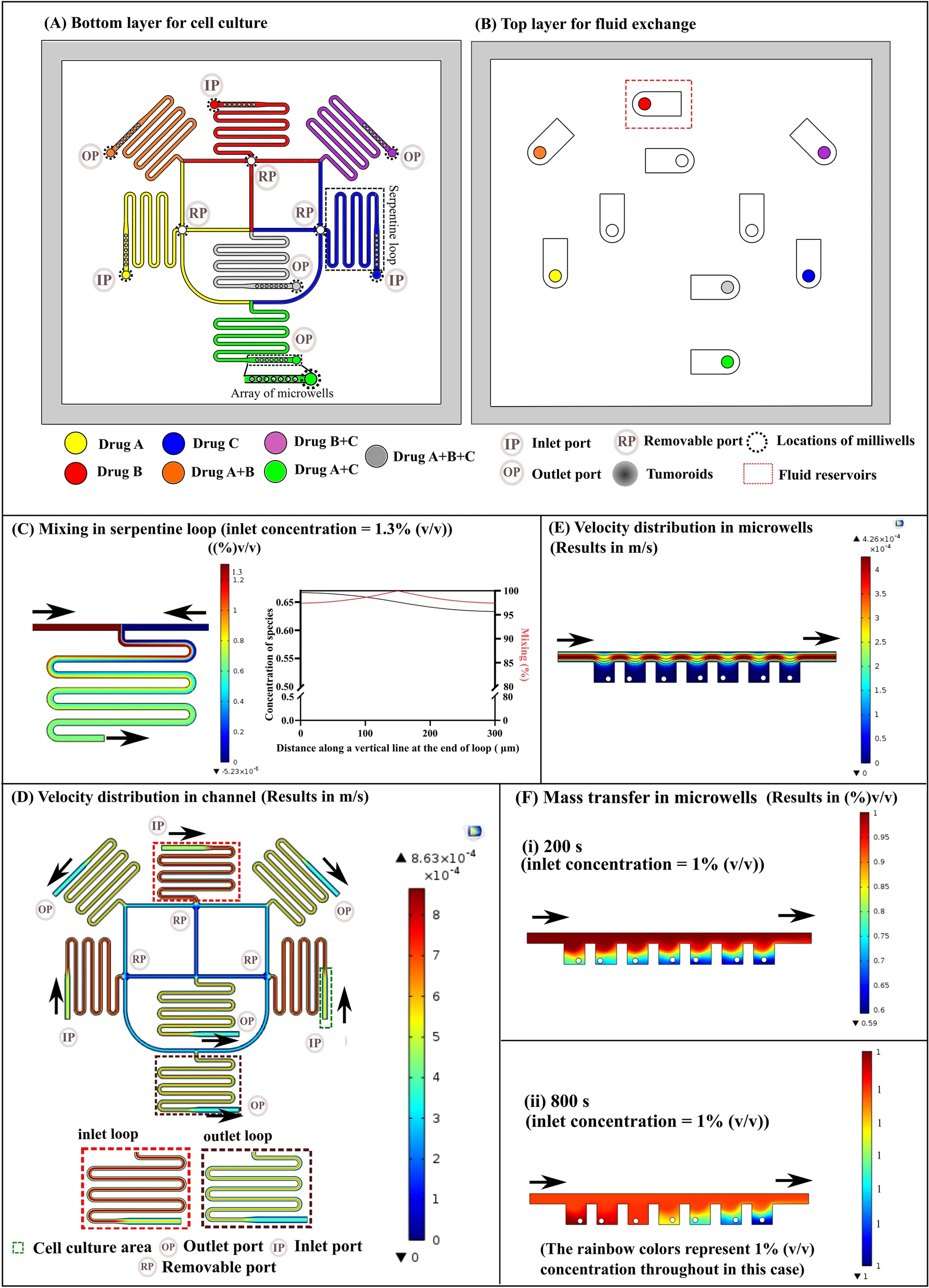
一种结合静电纺丝纳米纤维束的细胞球平台,用于评估水生生态毒理学。
Introduction
思路:
滥用化学品可以部分解释环境和人类健康问题发生率的增加。被称为生态毒理学的多学科领域研究环境污染物如何影响水生和陆地环境以及阐明生物体在接触污染物后的生理和生化反应。
在过去的几十年里,政府机构已经强制要求对活体动物进行毒性测试,以确定市场上新化学品或产品的环境风险。然而由于动物伦理方面的限制,需要合适的替代测试方案。
斑马鱼已成为水生生态毒理学中流行的替代模型系统。由于处于生命早期阶段的动物不属于明确的保护对象,因此斑马鱼胚胎具备环境和生物评估应用的可能性。
目前大部分体外试验都是基于细胞。研究表明,三维细胞培养物如细胞球相比于二维细胞培养物更能体现体内环境特征。 目前三维细胞球常用于哺乳动物细胞毒性测试,尚未广泛用于水生动物。
生物启发材料如生物启发的静电纺丝纳米纤维束(bioinspired strands of electrospun nanofibers, BSeNs)克服了传统细胞球系统的限制,并提供了诸如改善生存能力、增殖和氧气运输等优势。
本研究目的:
- 研究在斑马鱼肝细胞组成的三维细胞球中使用BSeN的效果;
- 通过基于细胞球的体外试验,研究BSeN在调节细胞球中细胞的活力、缺氧和功能方面的效果;
- 验证使用BSeN结合的三维细胞球和鱼胚胎急性毒性试验的相似性,研究潜在危险化学品和生殖毒性之间的相关性(FET,OECD TG 236)。
生物启发材料与水生环境中的细胞系结合起来,可以作为体外评估环境风险的平台。
Characterization of BSeNs
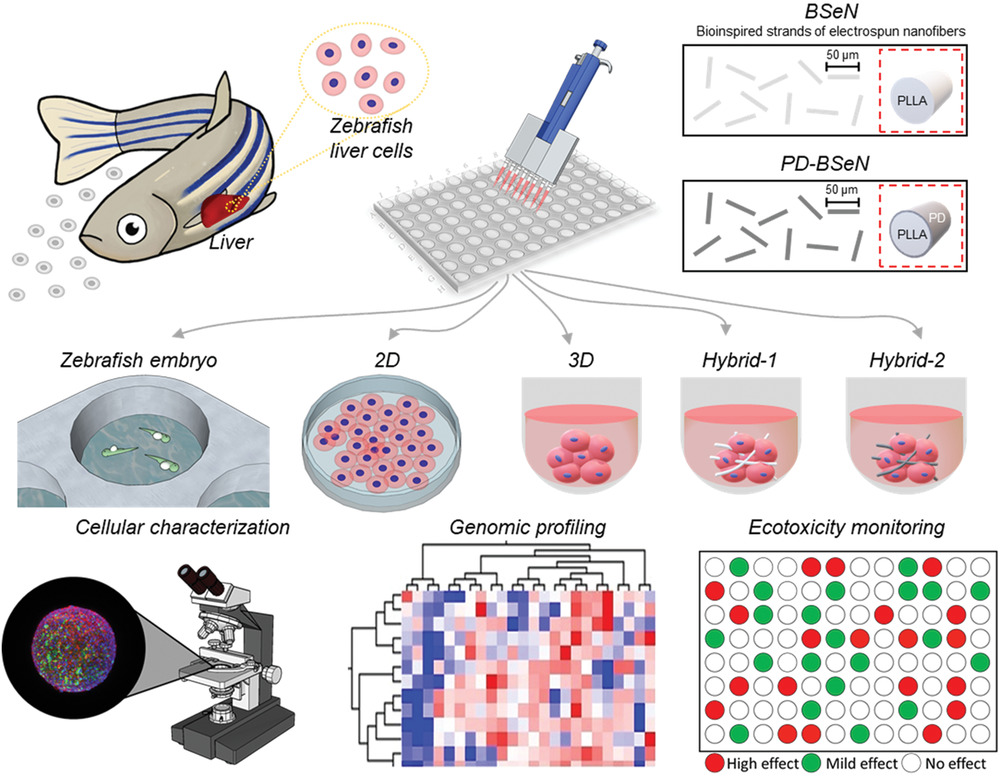
本研究的示意图。斑马鱼肝细胞被培养成单层、细胞球和混合细胞球。随后进行细胞特征分析、基因组分析和生态毒性监测应用。作者假设BSeN作为物理桥梁,能将氧气和营养物质运送到细胞球的核心,从而改善生存能力和增加增殖。
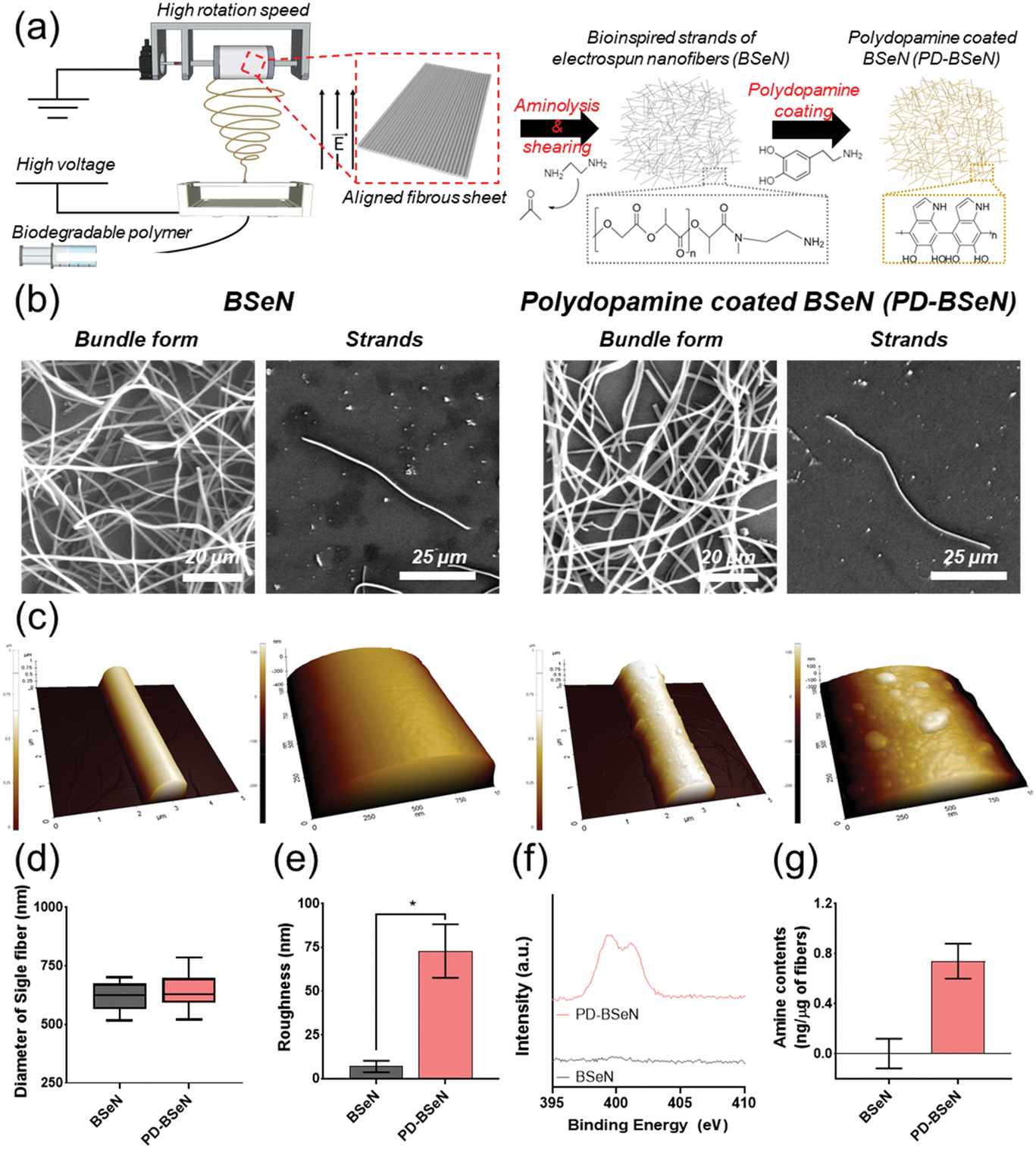
- (a)BSeN和PD-BSeN的生成示意图。通过裂解聚L-乳酸(PLLA)与乙二胺的酯键并产生具有酰胺键的短纤维,随后在表明覆盖聚多巴胺(Polydopamine,PD)以提高细胞亲和性。
- (b)BSeN和PD-BSeN的SEM图片。证实了纤维形式从网状排列变为分布良好、相对较小的短纤维。该尺寸与纤维蛋白相似,用于模仿ECM的纳米结构。
- (c)BSeN和PD-BSeN的AFM图片。图像显示,PD-BSeN更粗糙。
- (d)BSeN和PD-BSeN纤维的直径定量。
- (e)BSeN和PD-BSeN的表面粗糙度定量。
- (f)BSeN和PD-BSeN的X射线光电子能谱(XPS)。观察到清晰的氮峰(N1s)。
- (g)微BCA分析得到的BSeN和PD-BSeN的儿茶酚胺(Catecholamines)量化,因为多巴胺涂层是由于一个儿茶酚OH基去质子化而带负电,从而改善了亲水性和生物性能。
Effect of BSeN on 3D Spheroid Formation
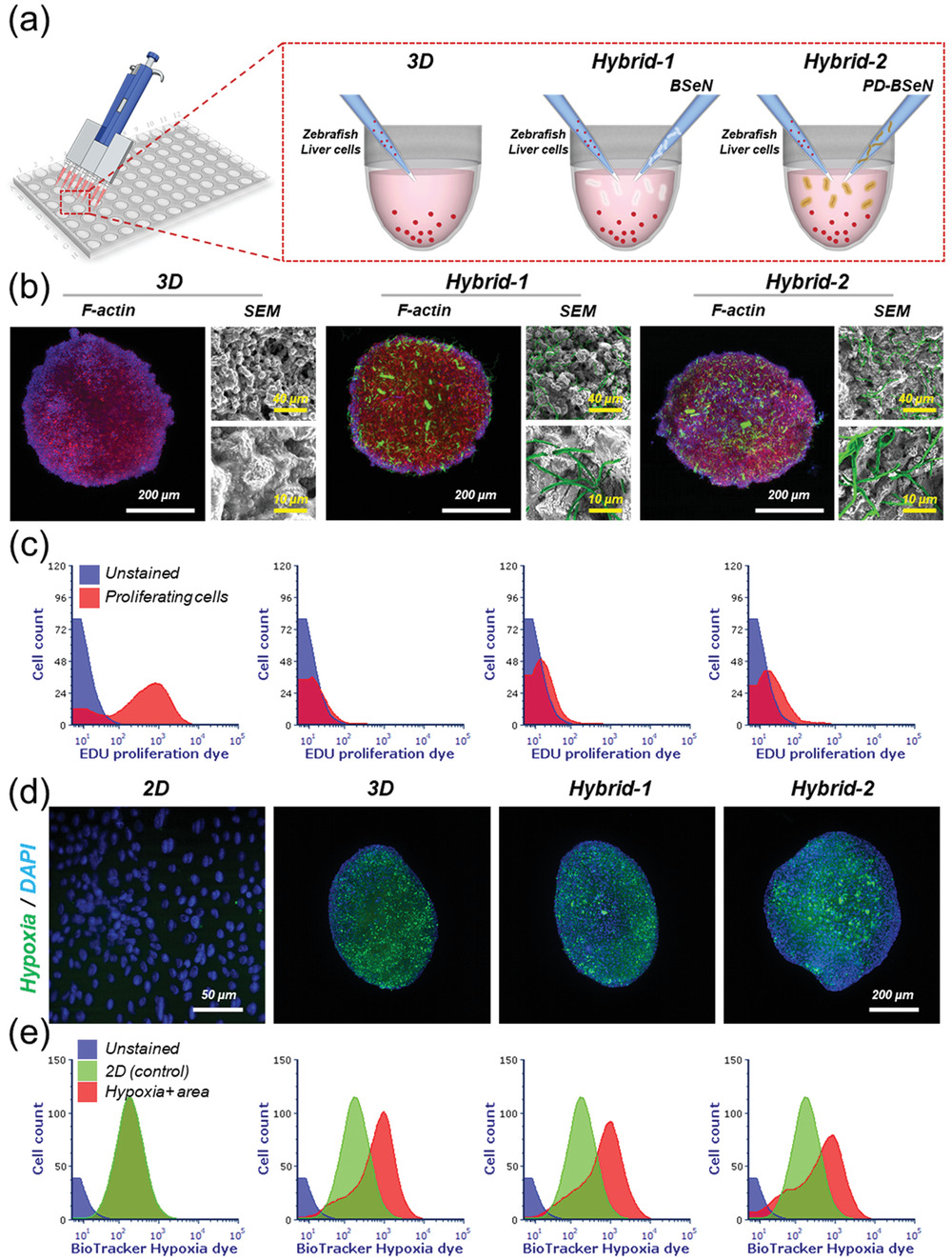
- (a) 用于制造细胞球(3D)、BSeN细胞球(Hybrid-1)和PD-BSeN细胞球(Hybrid-2)的方法示意图。
- (b)第3天细胞球的代表性细胞骨架结构和SEM显微图像。细胞核(DAPI,蓝色);F-肌动蛋白(红色);BSeN/PD-BSeN(FITC,绿色)。 证实了细胞球内均匀的细胞和纤维束的分布。
- (c)2D、3D、Hybrid-1和Hybrid-2组培养7天后细胞增殖的FACS结果。证实了BSeN可改善斑马鱼肝细胞的增殖。
- (d)第7天的代表性缺氧图像。细胞核(蓝色);缺氧(绿色)。
- (e)2D、3D、Hybrid-1和Hybrid-2组培养7天后缺氧的FACS结果。Hybrid-2组相比与3D组显示出缺氧减少。
Evaluation of Functions using Spheroid-Based In Vitro Assays
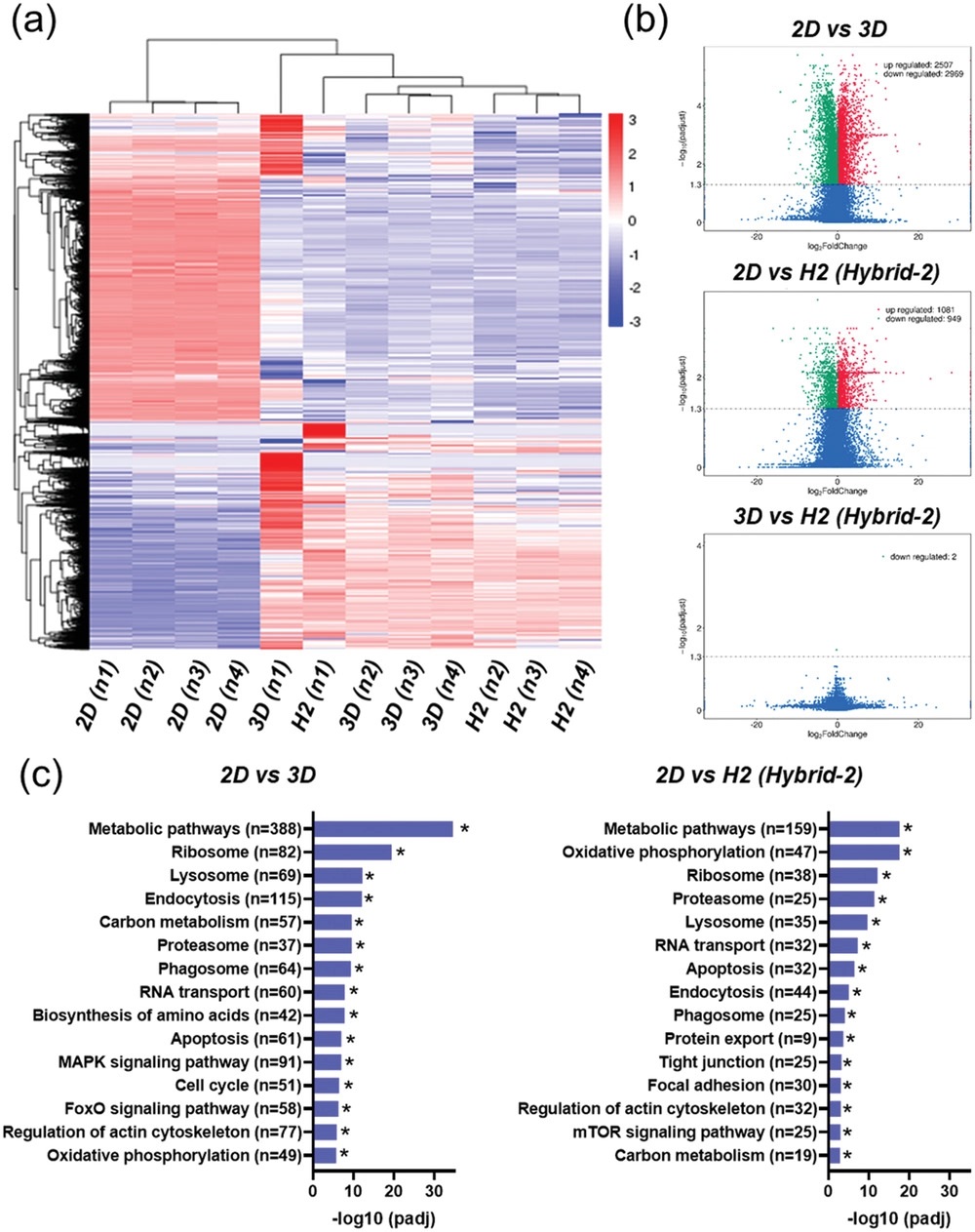
- (a)根据log2(FPKM+1)的热图层次聚类表示组中的差异表达基因(DEG)。红色和蓝色分别表示具有高和低表达水平的基因。分层聚类热图显示了2D和3D细胞球之间不同的表达谱。
- (b)火山图分析结果。图表示整体DEG分布。与2D组相比,3D细胞球表现出2507个基因的显著上调和2969个基因的下调。然而与3D组相比,Hybrid-2组未观察到明显差异。
- (c)转录本的KEGG富集直方图。3D和Hybrid-2组与代谢途径相关的基因丰度均高于2D组,Hybrid-2组的氧化磷酸化与2D组的氧化磷酸化显著不同,与PD-BSeN共存的细胞即使在氧气水平不足的情况下,也可以通过氧化磷酸化维持其增殖能力。

- (a)雌激素受体基因(ESR-1、ESR-β1和ESR-β2)的mRNA 表达倍数变化。与2D组相比,所有细胞球(3D、Hybrid-1和Hybrid-2)均表现出表达增加。
- (b)肝脏相关基因(HNF4A)的mRNA 表达倍数变化。2D组中的细胞显示出与3D组相似的HNF4A基因表达。然而,两个混合组都显示出比2D组更高的HNF4A基因表达。
- (c)药物代谢相关基因(GSTA1、PXR和CYP3C1)的mRNA 表达倍数变化。Hybrid-2组表现出比2D组更高的表达。
- (d)卵黄原蛋白相关基因(VTG-5)的mRNA表达倍数变化。VTG的相对基因表达在Hybrid-2组中高于2D、3D和Hybrid-1组。
- (e)卵黄蛋白原(VTG)的ELISA定量。发现即使培养14天,Hybrid-2组也显示出VTG蛋白表达稳定性。这些结果表明,3D培养方法可以改善细胞功能,但作为长期体外评估模型没有价值,除非它提供细胞可以长期存活的环境。
Comparison of Reproductive Toxicity in In Vitro Cell-Based Platforms and Zebrafish Embryos as a Result of EDC Exposure
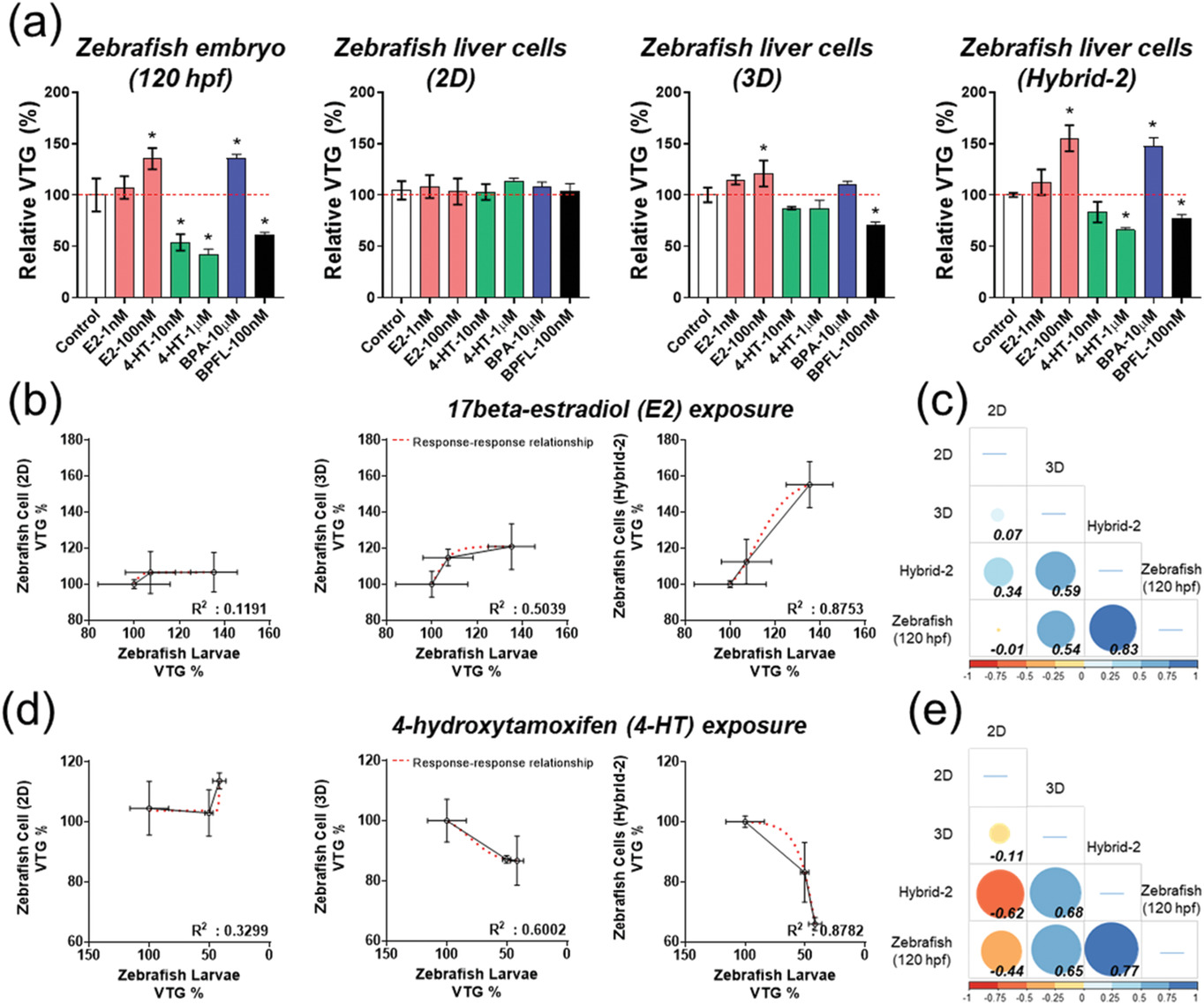
- (a)斑马鱼胚胎、2D、3D 和Hybrid-2组中的相对卵黄原蛋白(VTG)水平。发现相比2D组,3D组中VTG的趋势与使用斑马鱼胚胎观察到的趋势相似,而Hybrid-2组的结果最接近胚胎组。
- (b-c)17β-雌二醇(E2)暴露的相关矩阵图,计算组间的皮尔逊相关系数。
- (d-e)4-羟基三苯氧胺(4-HT)暴露的相关矩阵图,计算组间的皮尔逊相关系数。结果表明,使用具有集成仿生材料的3D细胞球平台获得的毒性数据与鱼胚胎急性毒性(Fish Embryo Acute Toxicity,FET)结果充分相关。
Conclusion
由于环境污染物会对各种分子机制产生不利影响,因此需要用于监测和功能研究的细胞模型作为生态毒理学中体内暴露研究的替代方案。本研究介绍了具有BSeN和水生细胞系的3D培养筛选平台,并研究了在环境毒性评估中替代动物试验方法的可能性。结果表明:
- BSeN的使用克服了传统细胞球系统带来的限制,并提供了诸如提高生存能力、增殖和氧气传输等优势;
- 体外试验分析表明,将BSeN与3D斑马鱼肝细胞培养物结合可以改善肝脏和生殖功能。
Reference
Park C G, Jun I, Lee S, et al. Integration of Bioinspired Fibrous Strands with 3D Spheroids for Environmental Hazard Monitoring[J]. Small, 2022.